Analysis of BioImages (in Progress)¶
This is becoming a collection of Python-based example codes focussing on image analysis of biological matter. Apologies for the code often being a little sloppy and "easy-going". Due to a limited time frame, priorities were set to "progress & visualization" instead of "finding the most efficient solution" :)
Whenever a function or method is developed to fulfill the goals, it is outsources into the subfolder "ModulesOwnAndExternalModified", take a look and have fun. Check out my website (www.bio-century.net) for more projects, literature recommendations, links to data bases and further inspirations.
# export image gallery yes or no
saveGallery = True
# saveGallery = False
Goal: Count nuclei of a fluorescence micrograph in an automated way
Steps 1: Import micrograph and compute a binary mask.
# sources
# - https://bbbc.broadinstitute.org/BBBC005
# - https://bbbc.broadinstitute.org/search/synthetic%20cells?
# - https://www.geeksforgeeks.org/distance-transformation-in-image-python-opencv/
# - https://www.geeksforgeeks.org/how-to-adjust-the-position-of-a-matplotlib-colorbar/
# - https://opencv24-python-tutorials.readthedocs.io/en/latest/py_tutorials/py_imgproc/py_watershed/py_watershed.html
# - https://scikit-image.org/docs/stable/auto_examples/segmentation/plot_watershed.html
# - https://stackoverflow.com/questions/70361424/extract-white-regions-from-binary-images
# - https://docs.scipy.org/doc/scipy/reference/generated/scipy.ndimage.binary_fill_holes.html
# - https://www.tutorialspoint.com/how-to-compute-the-area-and-perimeter-of-an-image-contour-using-opencv-python
# - https://www.youtube.com/watch?v=WQpXS9gBEu8
# data source
# Synthetic cells, Accession number BBBC005 · Version 1
# A. Lehmussola, P. Ruusuvuori, J. Selinummi, H. Huttunen and O. Yli-Harja,
# "Computational Framework for Simulating Fluorescence Microscope Images With Cell Populations,"
# in IEEE Transactions on Medical Imaging, vol. 26, no. 7, pp. 1010-1016, July 2007, doi: 10.1109/TMI.2007.896925.
#
# A. Lehmussola, P. Ruusuvuori, J. Selinummi, T. Rajala and O. Yli-Harja,
# "Synthetic Images of High-Throughput Microscopy for Validation of Image Analysis Methods,"
# in Proceedings of the IEEE, vol. 96, no. 8, pp. 1348-1360, Aug. 2008, doi: 10.1109/JPROC.2008.925490.
#
# - https://bbbc.broadinstitute.org/BBBC005
# - https://bbbc.broadinstitute.org/search/synthetic%20cells?
# data are published under CC0 1.0 Universal (CC0 1.0), https://creativecommons.org/publicdomain/zero/1.0/
# import modules
import cv2
from matplotlib import cm
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import make_axes_locatable
import numpy as np
from scipy import ndimage
# initialize and define variables and parameter
# fileName = "SIMCEPImages_A23_C96_F1_s19_w1.TIF" # mask
fileName = "SIMCEPImages_A23_C96_F1_s19_1_real.TIF" # "real" image
cellCountTruth = int(fileName[fileName.find('_C')+2:fileName.find('_F')]) # extract real cell number from filename
myThresholdValue = 70 # treshold value for binary mask
myImage = cv2.imread('./Data/' + fileName) # read image
myImageGray = cv2.cvtColor(myImage, cv2.COLOR_BGR2GRAY)
# ret, thresh = cv2.threshold(gray, 0, 1, cv2.THRESH_BINARY+cv2.THRESH_OTSU)
# fill holes
# define binary mask on computed on thresholded image
myImageThresholded = np.zeros(myImageGray.shape)
myImageThresholded[myImageGray > myThresholdValue] = 1
myImageThresholded = ndimage.binary_fill_holes(myImageThresholded).astype(int)
# plot figure
fig, ax = plt.subplots(figsize = (15, 7))
im = ax.imshow(myImageThresholded, cmap = cm.coolwarm)
divider = make_axes_locatable(ax)
cax = divider.new_vertical(size = "5%", pad = 0.3, pack_start = True)
fig.add_axes(cax)
cbar = fig.colorbar(im, cax = cax, ticks = [-1, 0, 1], orientation = 'horizontal')
TitleString = "\"Synthetic cells\": Binary Mask\n"
plt.title(TitleString, fontweight="bold", fontsize = 16, y = 22.3)
plt.suptitle("File: " + fileName, fontsize = 10, y = 0.91)
ax.set_xlabel("binary mask color scheme")
ax.set_xticks([])
ax.set_yticks([])
plt.show()
Step 2: Identify single objects and draw contours
# sources
# - https://www.kaggle.com/code/voglinio/separating-nuclei-masks-using-convexity-defects
# - https://stackoverflow.com/questions/32401806/get-mask-from-contour-with-opencv
# - https://stackoverflow.com/questions/50591442/convert-3-dim-image-array-to-2-dim
# - https://www.tutorialspoint.com/how-to-compute-the-area-and-perimeter-of-an-image-contour-using-opencv-python
# source of main algorithm
# - https://www.kaggle.com/code/voglinio/separating-nuclei-masks-using-convexity-defects
# User: Costas Voglis
# - https://de.mathworks.com/matlabcentral/answers/43435-i-couldn-t-understand-convex-area
# prop.convex_area / prop.filled_area
# convex_area (convex hull): the smallest region that satisfy two conditions: (1) it is convex (2) it contains
# the original region.
# => The ratio increases the less convex the area is (multiple nuclei)
# import modules
import cv2
import numpy as np
import matplotlib.pyplot as plt
from skimage.measure import regionprops
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
from statistics import mean
# initialize and define variables and parameter
# tresholds for
# 1Cell: single cell
# 2CellCluster: two cells
# gt2CellCluster: greater than two cells
dictThresholdValues = {}
dictThresholdValues["1Cell"] = 0
dictThresholdValues["2CellCluster"] = 1.065
dictThresholdValues["gt2CellCluster"] = 1.195
# compute results
# compute mean area for a single nucleus
meanFilledAreaTmp = Groundwork.meanFilledArea(myImage, myImageThresholded, dictThresholdValues)
# execute main algorith to identify cells and cell cluster
cellCount, contours, myImageContours, myMaskContoursAll = Groundwork.identifyCellContoursAndAreaOverlays(myImage,
myImageThresholded,
dictThresholdValues,
meanFilledAreaTmp)
# print table to compare cellcounts
print("-" * 27, end = '\n')
print(" Number of Cells")
print("-" * 27, end = '\n')
print("Detected: \t\t " + str(len(contours)))
print("True Value: \t\t " + str(cellCountTruth))
print("-" * 27, end = '\n')
print("Merged Cells Delta: \t" + str(len(contours) - cellCountTruth))
# plot figure
fig, ax = plt.subplots(figsize = (15, 7))
im = ax.imshow(myImageContours, cmap = cm.coolwarm)
divider = make_axes_locatable(ax)
cax = divider.new_vertical(size = "5%",
pad = 0.3,
pack_start = True)
TitleString = "\"Synthetic cells\": Determine Cell Contours\n"
plt.title(TitleString, fontweight = "bold", fontsize = 16)
plt.suptitle("File: " + fileName, fontsize = 10, y = 0.91)
ax.set_xticks([])
ax.set_yticks([])
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_01_01_CellContours.png', bbox_inches='tight')
plt.show()
---------------------------
Number of Cells
---------------------------
Detected: 74
True Value: 96
---------------------------
Merged Cells Delta: -22
For illustration purposes: Compute prop.convex_area-to-prop.filled_area distribution on the binary mask, which shall be taken as a criterium for multi-nuclei detection later on and defined in the above section.
# source of main algorithm
# - https://www.kaggle.com/code/voglinio/separating-nuclei-masks-using-convexity-defects
# User: Costas Voglis
# import modules
import cv2
import numpy as np
import matplotlib.pyplot as plt
from skimage.measure import regionprops
from statistics import mean
from matplotlib.gridspec import GridSpec
# contours, hierarchy = cv2.findContours(myImageThresholded.astype(np.uint8), 1, 2)
ratio_conv_filled_All = []
for ii, cnt in enumerate(contours):
M = cv2.moments(cnt)
area = cv2.contourArea(cnt)
perimeter = cv2.arcLength(cnt, True)
perimeter = round(perimeter, 1)
myImageContours = cv2.drawContours(myImage, [cnt], -1, (0, 0, 255), 2)
myMaskZeros = np.zeros(myImage.shape, np.uint8)
myMaskContoursTmp = cv2.drawContours(image = myMaskZeros, contours=[cnt], contourIdx = -1, color = (255, 255, 255), thickness = cv2.FILLED)
myMaskContours = np.zeros(myImage.shape)
myMaskContours[myMaskContoursTmp == 255] = 1
props = regionprops(myMaskContoursTmp, cache = False)
prop = props[0]
ratio_conv_filled_All.append(prop.convex_area / prop.filled_area)
# plot figure
fig = plt.figure(figsize = (16, 5))
gs = GridSpec(nrows = 1, ncols = 1)
ax0 = fig.add_subplot(gs[0, 0])
plt.hist(ratio_conv_filled_All, bins=30)
ax0 = plt.gca()
min_ylim, max_ylim = plt.ylim()
plt.axvline(dictThresholdValues["2CellCluster"], color='k', linestyle='dashed', linewidth = 1)
plt.text(dictThresholdValues["2CellCluster"]*1.005, max_ylim*0.9, 'Borderline 1 or 2 Cells: {:.2f}'.format(dictThresholdValues["2CellCluster"]))
plt.axvline(dictThresholdValues["gt2CellCluster"], color='k', linestyle='dashed', linewidth = 1)
plt.text(dictThresholdValues["gt2CellCluster"]*1.005, max_ylim*0.9, 'Borderline 2 or 3 Cells: {:.2f}'.format(dictThresholdValues["gt2CellCluster"]))
plt.grid(axis = 'y', linestyle = '--', linewidth = 0.3)
ax0.set_title('prop.convex_area / prop.filled_area')
ax0.set_xlabel('prop.convex_area / prop.filled_area')
ax0.set_ylabel('number of nucleic regions')
ax0.set_xticks(np.arange(1, 1.3, 0.01));
ax0.set_yticks(np.arange(0, 30, 1));
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_01_01_convex_vs_filled_area.png', bbox_inches='tight')
plt.show()
Step 3: Identify multi-nuclei regions
# sources
# - https://www.kaggle.com/code/voglinio/separating-nuclei-masks-using-convexity-defects
# - https://stackoverflow.com/questions/32401806/get-mask-from-contour-with-opencv
# - https://stackoverflow.com/questions/50591442/convert-3-dim-image-array-to-2-dim
# - https://www.tutorialspoint.com/how-to-compute-the-area-and-perimeter-of-an-image-contour-using-opencv-python
import warnings
warnings.filterwarnings('ignore')
# compute results
# print table to compare cellcounts
print("-" * 27, end = '\n')
print(" Number of Cells")
print("-" * 27, end = '\n')
print("Detected previously: \t " + str(len(contours)))
print("Detected now: \t\t " + str(cellCount))
print("True Value: \t\t " + str(cellCountTruth))
print("-" * 27, end = '\n')
print("Merged cells delta: \t " + str(cellCount - cellCountTruth))
print(myMaskContoursAll.shape)
print(np.max(myMaskContoursAll))
# plot figure
fig, ax = plt.subplots(figsize = (15, 7))
fig.set_figwidth(12)
fig.set_figheight(12)
ax.imshow(myMaskContoursAll/np.max(myMaskContoursAll))
TitleString = "\"Synthetic cells\": Detection of Multi-Core Overlays\n"
plt.title(TitleString, fontweight="bold", fontsize = 16)
plt.suptitle("File: " + fileName, fontsize = 10, y = 0.803)
ax.set_xticks([])
ax.set_yticks([])
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_01_01_Multicore_Overlays.png', bbox_inches='tight')
plt.show()
---------------------------
Number of Cells
---------------------------
Detected previously: 74
Detected now: 96
True Value: 96
---------------------------
Merged cells delta: 0
(520, 696, 3)
484.5
Goal: Count nuclei of a set of fluorescence micrographs in an automated way and identify the offset on each image by comparison to the ground truth number
# sources
# - https://www.kaggle.com/code/voglinio/separating-nuclei-masks-using-convexity-defects
# - https://pynative.com/python-list-files-in-a-directory/
# - https://stackoverflow.com/questions/32401806/get-mask-from-contour-with-opencv
# - https://stackoverflow.com/questions/50591442/convert-3-dim-image-array-to-2-dim
# - https://www.tutorialspoint.com/how-to-compute-the-area-and-perimeter-of-an-image-contour-using-opencv-python
# data source
# Synthetic cells, Accession number BBBC005 · Version 1
# A. Lehmussola, P. Ruusuvuori, J. Selinummi, H. Huttunen and O. Yli-Harja,
# "Computational Framework for Simulating Fluorescence Microscope Images With Cell Populations,"
# in IEEE Transactions on Medical Imaging, vol. 26, no. 7, pp. 1010-1016, July 2007, doi: 10.1109/TMI.2007.896925.
#
# A. Lehmussola, P. Ruusuvuori, J. Selinummi, T. Rajala and O. Yli-Harja,
# "Synthetic Images of High-Throughput Microscopy for Validation of Image Analysis Methods,"
# in Proceedings of the IEEE, vol. 96, no. 8, pp. 1348-1360, Aug. 2008, doi: 10.1109/JPROC.2008.925490.
# import modules
import cv2
import numpy as np
import matplotlib.pyplot as plt
import os
import os.path
from skimage.measure import regionprops
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
import matplotlib.pyplot as plt
import numpy as np
computeData = True
checkFiles = [os.path.exists('./Data/xpoints.npy') == True,
os.path.exists('./Data/ypoints.npy') == True]
if all(checkFiles):
xpoints = np.load('./Data/xpoints.npy', allow_pickle = True)
ypoints = np.load('./Data/ypoints.npy', allow_pickle = True)
computeData = False
if computeData == True:
xpoints = []
ypoints = []
# tresholds for
# 1Cell: single cell
# 2CellCluster: two cells
# gt2CellCluster: greater than two cells
dictThresholdValues = {}
dictThresholdValues["1Cell"] = 0
dictThresholdValues["2CellCluster"] = 1.05
dictThresholdValues["gt2CellCluster"] = 1.2
myThresholdValue = 70
meanFilledAreaAll = []
res = []
dir_path = './DataExternalSyntheticCells'
for path in os.listdir(dir_path):
if os.path.isfile(os.path.join(dir_path, path)):
res.append(path)
# compute mean area for a single nucleus with all file info
for ii, fileNameTmp in enumerate(res):
fileName = fileNameTmp
cellCountTruth = int(fileName[fileName.find('_C') + 2 : fileName.find('_F')])
myImage = cv2.imread('./DataExternalSyntheticCells/' + fileName)
myImageGray = cv2.cvtColor(myImage, cv2.COLOR_BGR2GRAY)
myImageThresholded = np.zeros(myImageGray.shape)
myImageThresholded[myImageGray > myThresholdValue] = 1
myImageThresholded = ndimage.binary_fill_holes(myImageThresholded).astype(int)
meanFilledArea = Groundwork.meanFilledArea(myImage, myImageThresholded, dictThresholdValues)
meanFilledAreaAll.append(meanFilledArea)
meanFilledArea = mean(meanFilledAreaAll)
# execute main algorith on all files to identify cells and cell cluster
print("-" * 138)
print(" " * 20 + "print scheme: " + "<filename> <TrueNumberNuclei> <ComputedNumberNuclei>")
print("-" * 138)
for ii, fileNameTmp in enumerate(res):
fileName = fileNameTmp
cellCountTruth = int(fileName[fileName.find('_C') + 2 : fileName.find('_F')])
myImage = cv2.imread('./DataExternalSyntheticCells/' + fileName)
myImageGray = cv2.cvtColor(myImage, cv2.COLOR_BGR2GRAY)
myImageThresholded = np.zeros(myImageGray.shape)
myImageThresholded[myImageGray > myThresholdValue] = 1
myImageThresholded = ndimage.binary_fill_holes(myImageThresholded).astype(int)
cellCount, contours, myImageContours, myMaskContoursAll = Groundwork.identifyCellContoursAndAreaOverlays(myImage,
myImageThresholded,
dictThresholdValues,
meanFilledArea)
if ii < 6:
print(fileName + ": " + str(cellCountTruth) + " " + str(cellCount) + "\t\t", end = '')
else:
print(fileName + ": " + str(cellCountTruth) + " " + str(cellCount) + "\t", end = '')
if (ii + 1) % 3 == 0:
print("\n", end = '')
xpoints.append(ii + 1)
ypoints.append(cellCountTruth - cellCount)
np.save('./Data/xpoints.npy', np.array(xpoints, dtype = object), allow_pickle = True)
np.save('./Data/ypoints.npy', np.array(ypoints, dtype = object), allow_pickle = True)
# plot figure
TitleString = "Quality Check: Automated Nuclei Detection"
x = np.linspace(0, 1, 2)
plt.plot(xpoints, ypoints)
plt.title(TitleString, fontweight="bold", fontsize = 16, y = 1.07)
plt.suptitle("File: " + fileName, fontsize = 10, y = 0.93)
plt.xlabel("File Index")
plt.ylabel("Difference [Truth - Detected_Cellcount]")
plt.xticks(np.arange(0, 72, 5))
plt.yticks(np.arange(0, 3, 1))
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_01_02_Quality_Auto_Nuclei_Detection.png', bbox_inches='tight')
plt.show()
1.1.3 Inspection of Curvature¶
Goal: Play with the curvature of the contour and see, if the results can be used for nuclei separation.
Step 1: Compute Curvature
# import modules
import cv2
from matplotlib import cm
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
from mpl_toolkits.axes_grid1 import make_axes_locatable
import numpy as np
from scipy import ndimage
# initialize and define variables and parameter
fileName = "test1.png"
myImage = cv2.imread('./Data/' + fileName)
myImageGray = cv2.cvtColor(myImage, cv2.COLOR_BGR2GRAY)
myImageThresholded = np.zeros(myImageGray.shape)
myImageThresholded[myImageGray > 0] = 1
myImageThresholded = ndimage.binary_fill_holes(myImageThresholded).astype(int)
# compute results
contours, hierarchy = cv2.findContours(myImageGray.astype(np.uint8), 1, 1)
for ii, cnt in enumerate(contours):
myMaskContoursAll2 = cv2.drawContours(myImage, [cnt], -1, (120,0,120), 1)
# plot figure
fig, ax = plt.subplots(figsize = (15, 7))
im = ax.imshow(myMaskContoursAll2)
TitleString = "Two Nuclei with Significant Bottleneck"
plt.title(TitleString, fontweight="bold", fontsize = 16, y = 1.01)
# plt.suptitle("File: " + fileName, fontsize = 10, y = 0.93)
ax.set_xticks([])
ax.set_yticks([])
plt.show()
# import modules
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
contoursxy = np.zeros((len(contours[0]), 2))
ll = list(contours[0])
contoursx = []
contoursy = []
# print(contoursx)
for ii in range(len(ll)):
contoursx.append(ll[ii][0][0])
contoursy.append(ll[ii][0][1])
contoursxy[:,0]=contoursx
contoursxy[:,1]=contoursy
# print(contoursxy)
contourCurvature, contourPixels = Groundwork.calculateContourCurvature(contoursxy)
print(contourCurvature[:,])
print(contourPixels)
print(max(contourCurvature))
[0.1767767 0.35777088 0.25 0.25 0.17888544 0. 0. 0.08944272 0.70710678 0.08944272 0. 0.17888544 0. 0. 0. 0.17888544 0.25 0.25 0.17888544 0.17888544 0.25 0. 0. 0. 0.25 0.17888544 0. 0. 0. 0. 0. 0.17888544 0.75 0.26516504 0.35777088 0.25 0.25 0.17888544 0.17888544 0.25 0. 0. 0.25 0.17888544 0.17888544 0. 0.17888544 0. 0.17888544 0. 0.08944272 0.70710678 0.08944272 0.17888544 0. 0.17888544 0.17888544 0.25 0. 0. 0. 0.25 0.17888544 0.17888544 0.25 0. 0. 0. 0. 0. 0. 0. 0.25 0.4472136 0.26832816 0.17888544 0. 0.17888544 0. 0.17888544 0.25 0. 0. 0. 0. 0.25 0.17888544 0.17888544 0.25 0. 0. 0. 0.25 0.17888544 0. 0.17888544 0.08838835 0.08838835 0.17888544 0.17888544 0. 0.17888544 0. 0.17888544 0.25 0. 0. 0.25 0.17888544 0.35777088 0.35777088 0.17888544 0. 0.17888544 0. 0. 0.17888544 0.5 0.35777088 0.1767767 0.35777088 0.25 0. 0. 0. 0. 0. 0. 0.25 0.17888544 0. 0.17888544 0.08838835 0.08838835 0.17888544 0.17888544 0.08838835 0.08838835 0.17888544 0. 0.17888544 0.25 0. 0. 0. 0. 0. 0.25 0.17888544 0.17888544 0.25 0.25 0.17888544 0. 0. 0. 0. 0. 0. 0. 0.26832816 0.26832816 0. 0.17888544 0. 0.17888544 0.17888544 0.25 0. 0. 0. 0. ] [[7, 33], [8, 32], [8, 31], [8, 30], [8, 29], [9, 28], [9, 27], [10, 26], [10, 25], [11, 25], [12, 24], [13, 23], [14, 23], [15, 22], [16, 22], [17, 21], [18, 21], [19, 21], [20, 21], [21, 20], [22, 20], [23, 20], [24, 20], [25, 20], [26, 20], [27, 20], [28, 21], [29, 21], [30, 22], [31, 22], [32, 23], [33, 23], [34, 24], [35, 23], [36, 22], [36, 21], [36, 20], [36, 19], [35, 18], [35, 17], [35, 16], [35, 15], [35, 14], [35, 13], [36, 12], [36, 11], [36, 10], [37, 9], [37, 8], [38, 7], [39, 6], [39, 5], [40, 5], [41, 4], [42, 4], [43, 4], [44, 3], [45, 3], [46, 3], [47, 3], [48, 3], [49, 3], [50, 3], [51, 4], [52, 4], [53, 4], [54, 4], [55, 4], [56, 4], [57, 4], [58, 4], [59, 4], [60, 4], [61, 4], [62, 5], [62, 6], [63, 7], [64, 8], [64, 9], [65, 10], [65, 11], [65, 12], [65, 13], [65, 14], [65, 15], [65, 16], [65, 17], [66, 18], [66, 19], [66, 20], [66, 21], [66, 22], [66, 23], [66, 24], [65, 25], [65, 26], [64, 27], [63, 28], [62, 29], [61, 29], [60, 30], [59, 31], [58, 31], [57, 32], [56, 32], [55, 32], [54, 32], [53, 32], [52, 32], [51, 33], [50, 33], [49, 32], [48, 32], [47, 32], [46, 31], [45, 31], [44, 30], [43, 30], [42, 30], [41, 31], [40, 32], [40, 33], [40, 34], [40, 35], [40, 36], [40, 37], [40, 38], [40, 39], [40, 40], [40, 41], [39, 42], [39, 43], [38, 44], [37, 45], [36, 46], [35, 46], [34, 47], [33, 48], [32, 49], [31, 49], [30, 50], [29, 50], [28, 50], [27, 50], [26, 50], [25, 50], [24, 50], [23, 50], [22, 50], [21, 49], [20, 49], [19, 49], [18, 49], [17, 48], [16, 48], [15, 47], [14, 47], [13, 46], [12, 46], [11, 45], [10, 45], [9, 44], [9, 43], [8, 42], [8, 41], [8, 40], [7, 39], [7, 38], [7, 37], [7, 36], [7, 35], [7, 34]] 0.75
Step 2: Plot Curvature and see, if there are significant maxima
# import modules
import numpy as np
import matplotlib.pyplot as plt
# compute results
print(len(contourCurvature))
y = contourCurvature
# plot figure
fig = plt.figure(figsize = (20, 5))
plt.plot(y)
plt.xticks(np.arange(0, len(contourCurvature), 5))
plt.axvline(32, color='k', linestyle='dashed', linewidth = 1)
plt.axvline(117, color='k', linestyle='dashed', linewidth = 1)
TitleString = "Curvature Value for Each Pixel of the Perimeter"
plt.title(TitleString, fontweight="bold", fontsize = 16, y = 1.01)
plt.xlabel("File Index")
plt.ylabel("Curvature Value")
plt.show()
print(contourPixels[28])
172
[30, 22]
Step 3: Filter with Gaussian in order to get reliable maxima detection
# import modules
import numpy as np
import matplotlib.pyplot as plt
from scipy.signal import savgol_filter
from scipy.ndimage import gaussian_filter
# compute results
print(len(contourCurvature))
y = contourCurvature
yhat = gaussian_filter(y, sigma=2)
# plot figure
fig = plt.figure(figsize = (20, 5))
plt.plot(yhat)
plt.xticks(np.arange(0, len(contourCurvature), 5))
plt.axvline(32, color='k', linestyle='dashed', linewidth = 1)
plt.axvline(117, color='k', linestyle='dashed', linewidth = 1)
TitleString = "Gauss-Filtered Curvature Value for Each Pixel of the Perimeter"
plt.title(TitleString, fontweight = "bold", fontsize = 16, y = 1.01)
plt.xlabel("File Index")
plt.ylabel("Gauss-Filtered Curvature Value")
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_01_03_Curvature_Colormap01.png', bbox_inches='tight')
plt.show()
print(contourPixels[28])
172
[30, 22]
Step 4: Find maxima on Gaussian-filtered curvature values
# import modules
from scipy.signal import find_peaks
# compute maxima
peaks, values = find_peaks(yhat, height = 0)
list2 = values['peak_heights']
list2.sort()
yhat1 = list(yhat)
xx1 = np.where(yhat == list2[-1])
xx2 = np.where(yhat == list2[-2])
# print results
print(int(xx1[0]))
print(int(xx2[0]))
33 118
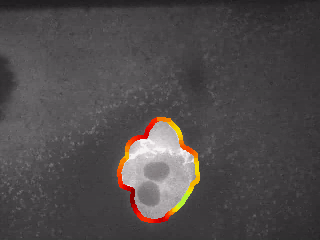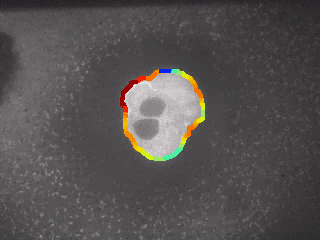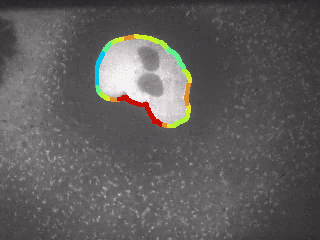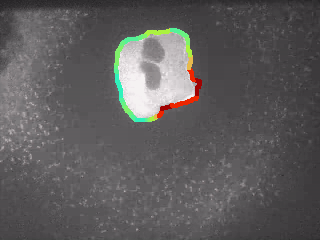
Step 5: Plot maxima of Gaussian-filtered curvature values into the image respecting their positions
# sources
# - https://en.wikipedia.org/wiki/Curvature
# source of algorithms
# - https://github.com/jmschabdach/caulobacter-curvature/blob/master/calculatingCellCurvature.py
myMaskContoursAllSepLine2 = myMaskContoursAll2
# plot figure
fig, ax = plt.subplots(figsize = (15, 7))
cv2.circle(myMaskContoursAllSepLine2, (contourPixels[int(xx1[0])][1], contourPixels[int(xx1[0])][0]), 2, (0, 120, 120), -1)
cv2.circle(myMaskContoursAllSepLine2, (contourPixels[int(xx2[0])][1], contourPixels[int(xx2[0])][0]), 2, (0, 120, 120), -1)
start_point = [contourPixels[int(xx1[0])][1], contourPixels[int(xx1[0])][0]]
end_point = [contourPixels[int(xx2[0])][1], contourPixels[int(xx2[0])][0]]
cv2.line(myMaskContoursAllSepLine2, start_point, end_point, (250, 0, 0), 2)
im = ax.imshow(myMaskContoursAllSepLine2)
divider = make_axes_locatable(ax)
cax = divider.new_vertical(size = "5%", pad = 0.3, pack_start = True)
TitleString = "\"Synthetic cells\": Binary Mask\n"
ax.set_xlabel("binary mask color scheme")
TitleString = "Cells with Curvature Bridge"
plt.title(TitleString, fontweight = "bold", fontsize = 14, y = 1.01)
plt.xlabel("x-position [px]")
plt.ylabel("y-position [px]")
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_01_03_Curvature_Colormap02.png', bbox_inches='tight')
plt.show()
Step 6: Plot Gaussian-filtered curvature values with jet-map color-coding
# import modules
import cv2
from matplotlib import cm
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
from mpl_toolkits.axes_grid1 import make_axes_locatable
import numpy as np
from scipy import ndimage
import warnings
warnings.filterwarnings('ignore')
# initialize and define variables and parameter
fileName = "test1.png"
myImage = cv2.imread('./Data/' + fileName)
myMaskContoursAllCurvatureValues = myImage
contoursxy[:,0]=contoursx
contoursxy[:,1]=contoursy
# print max values
print("-" * 27, end = '\n')
print(" Max Values")
print("-" * 27, end = '\n')
print(contourPixels[int(xx1[0])][1], contourPixels[int(xx1[0])][0])
print("-" * 27, end = '\n')
# compute results
myMaskContoursAllCurvatureValues = myImage
for ii in range(len(contourPixels)):
colorValue = 255*yhat[ii]/max(yhat)
colorPos = int(round(colorValue))
rcolor = int(round(cm.jet(colorPos)[0]*255))
gcolor = int(round(cm.jet(colorPos)[1]*255))
bcolor = int(round(cm.jet(colorPos)[2]*255))
rgbcolor = [rcolor, gcolor, bcolor]
# print(ii, contourCurvature[ii], colorPos, rgbcolor)
myMaskContoursAllCurvatureValues[int(contoursxy[ii,1])-1:int(contoursxy[ii,1])+1,int(contoursxy[ii,0])-1:int(contoursxy[ii,0])+1,:] = rgbcolor
# plot figure
fig, ax = plt.subplots(figsize = (15, 7))
im = ax.imshow(myMaskContoursAllCurvatureValues)
divider = make_axes_locatable(ax)
cax = divider.new_vertical(size = "5%", pad = 0.3, pack_start = True)
TitleString = "Cells with Color-Coded Curvature"
plt.title(TitleString, fontweight = "bold", fontsize = 14, y = 1.01)
plt.xlabel("x-position [px]")
plt.ylabel("y-position [px]")
cv2.circle(myMaskContoursAllCurvatureValues, (contourPixels[int(xx1[0])][1], contourPixels[int(xx1[0])][0]), 2, (0, 120, 120), -1)
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_01_03_Curvature_Colormap03.png', bbox_inches='tight')
plt.show()
---------------------------
Max Values
---------------------------
23 35
---------------------------
Goal: Ellipse fit of each single nuclei, multiple nuclei regions are to be excluded.
# sources
# - https://www.kaggle.com/code/voglinio/separating-nuclei-masks-using-convexity-defects
# - https://de.mathworks.com/matlabcentral/answers/43435-i-couldn-t-understand-convex-area
# - https://stackoverflow.com/questions/32401806/get-mask-from-contour-with-opencv
# - https://stackoverflow.com/questions/50591442/convert-3-dim-image-array-to-2-dim
# - https://stackoverflow.com/questions/62698756/opencv-calculating-orientation-angle-of-major-and-minor-axis-of-ellipse
# - https://www.tutorialspoint.com/how-to-compute-the-area-and-perimeter-of-an-image-contour-using-opencv-python
# - https://www.tutorialspoint.com/how-to-fit-the-ellipse-to-an-object-in-an-image-using-opencv-python
# sources of main algorithms
# https://www.kaggle.com/code/voglinio/separating-nuclei-masks-using-convexity-defects
# User: Costas Voglis
# and
# https://stackoverflow.com/questions/62698756/opencv-calculating-orientation-angle-of-major-and-minor-axis-of-ellipse
# User: fmw42, Fred Weinhaus
# import modules
import cv2
import numpy as np
import matplotlib.pyplot as plt
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
from skimage.measure import regionprops
import scipy.ndimage as ndimage
from mpl_toolkits.axes_grid1 import make_axes_locatable
fileName = "SIMCEPImages_A23_C96_F1_s02_w1.TIF"
cellCountTruth = int(fileName[fileName.find('_C')+2:fileName.find('_F')])
myThresholdValue = 70
myImage = cv2.imread('./DataExternalSyntheticCells/' + fileName)
myImageGray = cv2.cvtColor(myImage, cv2.COLOR_BGR2GRAY)
myImageThresholded = np.zeros(myImageGray.shape)
myImageThresholded[myImageGray > myThresholdValue] = 1
myImageThresholded = ndimage.binary_fill_holes(myImageThresholded).astype(int)
myImageEllipse = Groundwork.ellipseFit(myImage, myImageThresholded)
# plot figure
fig, ax = plt.subplots(figsize = (20, 10))
im = ax.imshow(myImageEllipse)
divider = make_axes_locatable(ax)
cax = divider.new_vertical(size = "5%",
pad = 0.3,
pack_start = True)
plt.title("Ellipse fit on nuclei", fontweight="bold", fontsize = 16, y = 1.05)
plt.suptitle("File: " + fileName, fontsize = 10, y = 0.91)
ax.set_xticks([])
ax.set_yticks([])
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_01_04_Ellipse_Fit_Nuclei.png', bbox_inches='tight')
plt.show()
Goal:
Analyse a first real fluorescence dataset and see how well the above algorithms work. This is really to play around with data and get a feeling of the algorithms, while there is no explicit scientific question posed here (due to a lack of data).
Plan:
- Step 1: Import fluorescence cell-based micrographs. To this end, we connect to a Image Data Resource-(idr-)-database and directly import a micrograph and its metadata via python
- Step 2: Preprocessing of the data received
- Step 3: Step 3: Preprocessing of data: Set up the masks, exclude debris
- Step 4: Apply ellipse fitting on nuclei and cytoplasm
Step 1: Import Fluorescence Cell-Based Micrographs and Metadata¶
Import and show data
# sources
# - https://idr.openmicroscopy.org/
# - https://biapol.github.io/blog/robert_haase/browsing_idr/readme.html
# - https://idr.openmicroscopy.org/webclient/api/plates/
# - https://matplotlib.org/stable/gallery/misc/table_demo.html#sphx-glr-gallery-misc-table-demo-py
# - https://matplotlib.org/stable/gallery/subplots_axes_and_figures/subfigures.html
# - https://stackoverflow.com/questions/31726643/how-to-plot-in-multiple-subplots
# - https://towardsdatascience.com/simple-little-tables-with-matplotlib-9780ef5d0bc4
# - https://towardsdatascience.com/plot-organization-in-matplotlib-your-one-stop-guide-if-you-are-reading-this-it-is-probably-f79c2dcbc801
# data source
# https://idr.openmicroscopy.org/webclient/api/plates/
# https://idr.openmicroscopy.org/webclient/img_detail/3232695/
# 41744_illum_corrected [Well G9, Field 7]
# data are published under CC BY 4.0, https://creativecommons.org/licenses/by/4.0/
# import modules
import json
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import make_axes_locatable
import numpy as np
import requests
from skimage.io import imread, imshow
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
import numpy as np
# initialize and define variables and parameter
screen_id = 1751
imageID = "3232695"
myLink = "https://idr.openmicroscopy.org/webclient/render_image/" + imageID + "/"
dictImageMetaData = {}
cell_text = []
# load data
################ Import metadata ################
# configure which dataset to browse
# screen_id = 2302
MAP_URL = "https://idr.openmicroscopy.org/webclient/api/annotations/?type=map&{type}={screen_id}"
# open an interface to the internet
with requests.Session() as session:
# turn the generic MAP_URL into a specific URL for the screen
qs = {'type': 'screen', 'screen_id': screen_id}
qs = {'image_id': imageID}
IMAGE_DETAILS_URL = "https://idr.openmicroscopy.org/webclient/imgData/{image_id}/"
url = IMAGE_DETAILS_URL.format(**qs)
r = session.get(url)
with open("./Data/02_01_ImageMetaData.json", "w") as outfile:
if r.status_code == 200:
json.dump(r.json(), outfile, indent = 4, sort_keys = True)
with open('./Data/02_01_ImageMetaData.json', 'r') as openfile:
dictImageMetaData = json.load(openfile)
# compute results
################ Import image ################
myImage = imread(myLink)
# table 1: file imformation
tableColumnsFileInfo = ["File Info"]
tableRowsFileInfo = ['myLink', 'MAP_URL', 'IMAGE_DETAILS_URL', 'imageId', 'tiles', 'wellId', 'height', 'width']
tableDataFileInfo = [
[str(myLink)],
[str(MAP_URL)],
[str(url)],
[str(dictImageMetaData["meta"]['imageId'])],
[str(dictImageMetaData["tiles"])],
[str(dictImageMetaData["meta"]['wellId'])],
[str(dictImageMetaData["size"]['height'])],
[str(dictImageMetaData["size"]['width'])],
]
cell_text0 = []
for row in range(len(tableDataFileInfo)):
cell_text0.append(tableDataFileInfo[row])
# table 2: channel information
tableColumnsChannelInfo = ['label', 'active status', 'hexacode']
tableRowsChannelInfo = ['Red Channel','Green Channel','Blue Channel','Blue Channel','Blue Channel']
tableDataChannelInfo = [
[str(dictImageMetaData["channels"][0]['label']), str(dictImageMetaData["channels"][0]['active']), str(dictImageMetaData["channels"][0]['color'])],
[str(dictImageMetaData["channels"][1]['label']), str(dictImageMetaData["channels"][1]['active']), str(dictImageMetaData["channels"][1]['color'])],
[str(dictImageMetaData["channels"][2]['label']), str(dictImageMetaData["channels"][2]['active']), str(dictImageMetaData["channels"][2]['color'])],
[str(dictImageMetaData["channels"][3]['label']), str(dictImageMetaData["channels"][3]['active']), str(dictImageMetaData["channels"][3]['color'])],
[str(dictImageMetaData["channels"][4]['label']), str(dictImageMetaData["channels"][4]['active']), str(dictImageMetaData["channels"][4]['color'])],
]
# there is no pixel size available for this micrograph!
cell_text = []
for row in range(len(tableDataChannelInfo)):
cell_text.append(tableDataChannelInfo[row])
rowColors = [
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
]
colColors = [
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
]
colColors0 = [
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
]
rowColors0 = [
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
[0.54901961, 0.58559016, 0.77517878, 1. ],
]
################ show figure ################
# set up figure grid
fig = plt.figure(figsize = (20, 10))
gs = GridSpec(nrows = 1, ncols = 1)
# plot image
ax0 = fig.add_subplot(gs[0, 0])
ax0.imshow(myImage)
# ax0 = plt.gca()
ax0.get_xaxis().set_visible(False)
ax0.get_yaxis().set_visible(False)# Hide axes border
plt.suptitle("Fluorescence Data", fontweight = "bold", fontsize = 16, y = 0.9)
plt.draw()
plt.show()
fig = plt.figure(figsize = (10, 4.5))
gs = GridSpec(nrows = 2, ncols = 1)
ax1 = fig.add_subplot(gs[0, 0])
the_table = plt.table(cellText = cell_text0,
rowLabels = tableRowsFileInfo,
rowColours = rowColors0,
colLabels = tableColumnsFileInfo,
colColours = colColors0,
loc = 'center')
the_table.scale(0.9, 1.4)
ax1 = plt.gca()
ax1.get_xaxis().set_visible(False)
ax1.get_yaxis().set_visible(False)
plt.box(on = None)
ax2 = fig.add_subplot(gs[1, 0])
the_table = plt.table(cellText = cell_text,
rowLabels = tableRowsChannelInfo,
rowColours = rowColors,
colLabels = tableColumnsChannelInfo,
colColours = colColors,
loc = 'center')
the_table.scale(0.9, 1.4)
ax2 = plt.gca()
ax2.get_xaxis().set_visible(False)
ax2.get_yaxis().set_visible(False)
plt.box(on = None)
fig = plt.figure(figsize = (20, 7))
gs = GridSpec(nrows = 1, ncols = 3)
ax3 = fig.add_subplot(gs[0, 0])
plt.imshow(myImage[:,:,0])
ax3 = plt.gca()
ax3.set_title('Hoechst')
ax3.get_xaxis().set_visible(False)
ax3.get_yaxis().set_visible(False)
ax4 = fig.add_subplot(gs[0, 1])
plt.imshow(myImage[:,:,1])
ax4 = plt.gca()
ax4.set_title('ERSyto')
ax4.get_xaxis().set_visible(False)
ax4.get_yaxis().set_visible(False)
ax5 = fig.add_subplot(gs[0, 2])
plt.imshow(myImage[:,:,2])
ax5 = plt.gca()
ax5.set_title('ERSytoBleed')
ax5.get_xaxis().set_visible(False)
ax5.get_yaxis().set_visible(False)
Step 2: Save image¶
# import modules
import cv2
# save a image using extension
cv2.imwrite("./Data/02_01_myImageRGB.tif", myImage)
cv2.imwrite("./Data/02_01_myImageR.tif", myImage[:,:,0])
cv2.imwrite("./Data/02_01_myImageG.tif", myImage[:,:,1])
cv2.imwrite("./Data/02_01_myImageB.tif", myImage[:,:,2])
True
Step 3: Preprocessing of data: Set up the masks, exclude debris¶
Define nuclei mask
# import modules
import cv2
from matplotlib import cm
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import make_axes_locatable
import numpy as np
from scipy import ndimage
# define filenames and parameters
fileName = "02_01_myImageR.tif"
myThresholdValue = 30
myImage = cv2.imread('./Data/' + fileName)
myImageGray = cv2.cvtColor(myImage, cv2.COLOR_BGR2GRAY)
# ret, thresh = cv2.threshold(gray, 0, 1, cv2.THRESH_BINARY + cv2.THRESH_OTSU)
# fill holes
# define binary mask on computed on thresholded image
myImageThresholded = np.zeros(myImageGray.shape)
myImageThresholded[myImageGray > myThresholdValue] = 1
myImageThresholded = ndimage.binary_fill_holes(myImageThresholded).astype(int)
# plot figure
fig = plt.figure(figsize = (20, 10))
gs = GridSpec(nrows = 1, ncols = 3)
ax0 = fig.add_subplot(gs[0, 0])
plt.imshow(myImage[:,:,0])
ax0 = plt.gca()
ax0.set_title('Hoechst')
ax0.get_xaxis().set_visible(False)
ax0.get_yaxis().set_visible(False)
ax1 = fig.add_subplot(gs[0, 1])
plt.imshow(myImageThresholded)
ax1 = plt.gca()
ax1.set_title('Nuclei Binary Mask')
ax1.get_xaxis().set_visible(False)
ax1.get_yaxis().set_visible(False)
Remove small objects from nuclei mask
from skimage import morphology
# sources
# - https://scikit-image.org/docs/dev/api/skimage.morphology.html#skimage.morphology.remove_small_objects
# remove small objects
minimum_pixelarea = 30
myImageThresholdedtmp = []
myImageThresholdedrso = []
myImageThresholdedtmp = myImageThresholded
myImageThresholdedtmp[980+20:1030, 550:585] = 0
myImageThresholdedtmp[980:1025, 550+25:585+20] = 0
myImageThresholdedtmp[990+2:1010, 550:585+20] = 0
myImageThresholdedrso = morphology.remove_small_objects(np.array(myImageThresholdedtmp, dtype=bool), minimum_pixelarea)
myImageRThresholdedRSO = myImageThresholdedrso
# plot figure
fig, ax = plt.subplots(figsize = (15, 7))
im = ax.imshow(myImageThresholdedrso, cmap = cm.coolwarm)
divider = make_axes_locatable(ax)
cax = divider.new_vertical(size = "5%", pad = 0.3, pack_start = True)
fig.add_axes(cax)
cbar = fig.colorbar(im, cax = cax, ticks = [-1, 0, 1], orientation = 'horizontal')
plt.suptitle("Nuclei Binary Mask Cleanup", fontweight = "bold", fontsize = 16, y = 0.92)# Add footer
ax.set_xlabel("binary mask color scheme")
# ax.set_xticks([])
# ax.set_yticks([])
plt.show()
Define mask for cytoplasm and mask out debris
# sources
# - https://scikit-image.org/docs/dev/api/skimage.morphology.html#skimage.morphology.remove_small_objects
# import modules
from skimage import morphology
# define filenames and parameters
fileName = "02_01_myImageB.tif"
myThresholdValue = 10
myImage = cv2.imread('./Data/' + fileName)
myImageGray = cv2.cvtColor(myImage, cv2.COLOR_BGR2GRAY)
# ret, thresh = cv2.threshold(gray, 0, 1, cv2.THRESH_BINARY + cv2.THRESH_OTSU)
# fill holes
# define binary mask on computed on thresholded image
myImageThresholded = np.zeros(myImageGray.shape)
myImageThresholded[myImageGray > myThresholdValue] = 1
myImageThresholded = ndimage.binary_fill_holes(myImageThresholded).astype(int)
# remove small objects
minimum_pixelarea = 30
myImageThresholdedtmp = []
myImageThresholdedrso = []
myImageThresholdedtmp = myImageThresholded
# remove specific debris
myImageThresholdedtmp[980+20:1030, 550:585] = 0
myImageThresholdedtmp[980:1025, 550+25:585 + 20] = 0
myImageThresholdedtmp[990+2:1010, 550:585 + 20] = 0
myImageThresholdedrso = morphology.remove_small_objects(np.array(myImageThresholdedtmp, dtype = bool), minimum_pixelarea)
myImageGThresholdedRSO = myImageThresholdedrso
# plot figure
fig = plt.figure(figsize = (20, 10))
gs = GridSpec(nrows = 1, ncols = 3)
ax0 = fig.add_subplot(gs[0, 0])
plt.imshow(myImage[:,:,0]*2)
ax0 = plt.gca()
ax0.set_title('Cytoplasm')
ax0.get_xaxis().set_visible(False)
ax0.get_yaxis().set_visible(False)
ax1 = fig.add_subplot(gs[0, 1])
plt.imshow(myImageThresholdedrso)
ax1 = plt.gca()
ax1.set_title('Cytoplasm Binary Mask')
ax1.get_xaxis().set_visible(False)
ax1.get_yaxis().set_visible(False)
Review masks
# compute results
contours, myImageContours, myMaskRContoursAll, myMaskRContoursAllLabel = Groundwork.identifyCellContours(myImage, myImageRThresholdedRSO, showCenterOfMass=False, perimeterColor=[20, 240, 20])
contours, myImageContours, myMaskGContoursAll, myMaskGContoursAllLabel = Groundwork.identifyCellContours(myImage, myImageGThresholdedRSO, showCenterOfMass=False, perimeterColor=[20, 240, 20])
contours, myImageContours, myMaskBContoursAll, myMaskBContoursAllLabel = Groundwork.identifyCellContours(myImage, myImageGThresholdedRSO, showCenterOfMass=False, perimeterColor=[20, 20, 240])
myMaskContoursAll = np.zeros(myImage.shape)
myMaskContoursAll[myMaskGContoursAll==1] = 100
myMaskContoursAll[myMaskRContoursAll==1] = 255
print(np.min(myMaskGContoursAllLabel))
print(np.max(myMaskGContoursAllLabel))
# plot figure
fig = plt.figure(figsize = (20, 19))
gs = GridSpec(nrows = 1, ncols = 3)
ax0 = fig.add_subplot(gs[0, 0])
plt.imshow(myImageContours*2)
ax0 = plt.gca()
ax0.set_title('Cells With Boundaries')
ax0.get_xaxis().set_visible(False)
ax0.get_yaxis().set_visible(False)
ax1 = fig.add_subplot(gs[0, 1])
plt.imshow(myMaskContoursAll.astype(np.uint8))
ax1 = plt.gca()
ax1.set_title('Cell Binary Masks')
ax1.get_xaxis().set_visible(False)
ax1.get_yaxis().set_visible(False)
ax2 = fig.add_subplot(gs[0, 2])
plt.imshow(myMaskGContoursAllLabel.astype(np.uint8))
ax2 = plt.gca()
ax2.set_title('Cell Binary Masks with Labels')
ax2.get_xaxis().set_visible(False)
ax2.get_yaxis().set_visible(False)
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_02_02_Cells_With_Boundaries.png', bbox_inches='tight')
plt.show()
0.0 78.0
Eliminate everything without a nucleus from cytoplasmic mask
# import modules
import math
# define filenames and parameters
plotstatus = False
jj=0
myMaskContoursAllCorrected = np.zeros((myImage.shape[0], myImage.shape[1],int(np.max(myMaskBContoursAllLabel))))
print(myMaskContoursAllCorrected.shape)
# compute results
for ii in range(1,int(np.max(myMaskBContoursAllLabel))+1):
myMaskTmp = np.zeros((myImage.shape[0], myImage.shape[1]))
myMaskTmp[myMaskBContoursAllLabel==ii]=1
if np.max(myImageRThresholdedRSO + myMaskTmp) > 1:
print(str(ii) + ", ", end='')
myMaskContoursAllCorrected[:,:,ii-1] = myMaskTmp
# plot figure
if plotstatus==True:
f, axarr = plt.subplots(20,5,figsize = (20, 100))
for ii in range(int(np.max(myMaskBContoursAllLabel))):
xindex = ii % 5
yindex = math.floor(ii / 5)
axarr[yindex,xindex].imshow(myMaskContoursAllCorrected[:,:,ii])
axarr[yindex,xindex].set_title(ii)
(1080, 1080, 78) 3, 4, 6, 7, 8, 9, 10, 11, 12, 14, 15, 17, 18, 20, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 34, 35, 36, 37, 39, 41, 42, 43, 44, 45, 46, 47, 49, 50, 52, 53, 54, 55, 56, 57, 58, 59, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 76, 77, 78,
Review masks
# compute results
result = np.sum(myMaskContoursAllCorrected[:,:,1:], axis = 2)
myMaskContoursAll_Marks = cv2.imread('./Figures/myMaskContoursAll_Marks.png')
# plot figure
fig = plt.figure(figsize = (16, 15))
gs = GridSpec(nrows = 2, ncols = 2)
ax0 = fig.add_subplot(gs[0, 0])
plt.imshow(myMaskContoursAll_Marks.astype(np.uint8))
ax0 = plt.gca()
ax0.set_title('Cell Binary Masks')
ax0.get_xaxis().set_visible(False)
ax0.get_yaxis().set_visible(False)
cv2.imwrite('./Figures/myMaskContoursAll.png', myMaskContoursAll)
ax1 = fig.add_subplot(gs[0, 1])
plt.imshow(result)
ax1 = plt.gca()
ax1.set_title('Cell Binary Masks after Cleanup')
ax1.get_xaxis().set_visible(False)
ax1.get_yaxis().set_visible(False)
Ellipse fit on nuclei and whole cell
# sources
# - https://www.kaggle.com/code/voglinio/separating-nuclei-masks-using-convexity-defects
# - https://de.mathworks.com/matlabcentral/answers/43435-i-couldn-t-understand-convex-area
# - https://stackoverflow.com/questions/32401806/get-mask-from-contour-with-opencv
# - https://stackoverflow.com/questions/50591442/convert-3-dim-image-array-to-2-dim
# - https://stackoverflow.com/questions/62698756/opencv-calculating-orientation-angle-of-major-and-minor-axis-of-ellipse
# - https://www.tutorialspoint.com/how-to-compute-the-area-and-perimeter-of-an-image-contour-using-opencv-python
# - https://www.tutorialspoint.com/how-to-fit-the-ellipse-to-an-object-in-an-image-using-opencv-python
# sources of main algorithms
# https://www.kaggle.com/code/voglinio/separating-nuclei-masks-using-convexity-defects
# User: Costas Voglis
# and
# https://stackoverflow.com/questions/62698756/opencv-calculating-orientation-angle-of-major-and-minor-axis-of-ellipse
# User: fmw42, Fred Weinhaus
# import modules
import cv2
import numpy as np
import matplotlib.pyplot as plt
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
from skimage.measure import regionprops
import math
fileName = "02_01_myImageRGB.tif"
myImage = cv2.imread('./Data/' + fileName)
# compute results
myImageEllipseR = Groundwork.ellipseFit(myMaskContoursAll.astype(np.uint8),
myImageRThresholdedRSO, 4000)
# plot figure
fig = plt.figure(figsize = (16, 15))
gs = GridSpec(nrows = 1, ncols = 1)
ax0 = fig.add_subplot(gs[0, 0])
plt.imshow(myImageEllipseR)
ax0 = plt.gca()
ax0.set_title('Ellipse Fit on Nucei')
ax0.get_xaxis().set_visible(False)
ax0.get_yaxis().set_visible(False)
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_02_03_Ellipse_Fit_Real_Cells.png', bbox_inches='tight')
plt.show()
Compute Mean Area of Nuclei and Mean Intensity
# import modules
import math
# define filenames and parameters
plotstatus = False
jj = 0
myMaskRContoursAllCorrected = np.zeros((myImage.shape[0], myImage.shape[1],int(np.max(myMaskRContoursAllLabel))))
myMaskTmpIntensity = np.zeros((myImage.shape[0], myImage.shape[1],int(np.max(myMaskRContoursAllLabel))))
filledAreaNuclei = []
meanIntensityNuclei = []
print(myMaskContoursAllCorrected.shape)
# compute results
for ii in range(1,int(np.max(myMaskRContoursAllLabel))+1):
myMaskTmp = np.zeros((myImage.shape[0], myImage.shape[1]))
myMaskTmp[myMaskRContoursAllLabel==ii]=1
if np.max(myImageRThresholdedRSO + myMaskTmp) > 1:
print(str(ii) + ", ", end = '')
myMaskRContoursAllCorrected[:,:,ii-1] = myMaskTmp
numberOfPixel = np.sum(np.sum(myMaskTmp, axis = 0), axis = 0)
filledAreaNuclei.append(numberOfPixel)
myMaskTmpIntensity[:,:,ii-1]=myMaskTmp*myImage[:,:,2]
integratedIntensity = np.sum(np.sum(myMaskTmp*myImage[:,:,2], axis = 0), axis = 0)
meanIntensityNuclei.append(integratedIntensity / numberOfPixel)
# plot figure
if plotstatus==True:
f, axarr = plt.subplots(17,5,figsize = (20, 100))
for ii in range(int(np.max(myMaskRContoursAllLabel))):
xindex = ii % 5
yindex = math.floor(ii / 5)
axarr[yindex,xindex].imshow(myMaskTmpIntensity[:,:,ii])
axarr[yindex,xindex].set_title(ii)
print("")
print("filledAreaNuclei", filledAreaNuclei)
print("meanIntensityNuclei", meanIntensityNuclei)
(1080, 1080, 78) 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, filledAreaNuclei [501.0, 683.0, 1072.0, 936.0, 988.0, 913.0, 739.0, 778.0, 1035.0, 1056.0, 666.0, 705.0, 713.0, 838.0, 1016.0, 927.0, 1020.0, 886.0, 747.0, 862.0, 801.0, 1012.0, 770.0, 1169.0, 775.0, 775.0, 1125.0, 714.0, 786.0, 247.0, 873.0, 1059.0, 1230.0, 1076.0, 967.0, 749.0, 669.0, 954.0, 1002.0, 883.0, 664.0, 30.0, 769.0, 835.0, 1134.0, 914.0, 907.0, 671.0, 757.0, 592.0, 678.0, 868.0, 780.0, 777.0, 636.0, 641.0, 873.0, 341.0, 767.0, 648.0, 706.0, 629.0, 470.0, 843.0, 883.0, 1062.0, 608.0, 733.0, 1447.0, 386.0, 243.0, 766.0, 1103.0, 914.0, 967.0, 110.0, 910.0, 1105.0, 638.0, 812.0, 1076.0, 614.0, 173.0, 390.0] meanIntensityNuclei [27.744510978043913, 31.275256222547583, 30.090485074626866, 35.36645299145299, 31.772267206477732, 30.428258488499452, 25.882273342354534, 29.443444730077122, 33.19420289855073, 30.334280303030305, 31.06756756756757, 31.78581560283688, 33.92145862552594, 33.582338902147974, 36.56496062992126, 47.49514563106796, 35.19803921568627, 32.20428893905192, 30.97590361445783, 29.858468677494198, 35.90761548064919, 41.30335968379447, 27.41948051948052, 35.221556886227546, 28.032258064516128, 29.52516129032258, 31.232, 28.84173669467787, 29.0381679389313, 25.453441295546558, 31.079037800687285, 32.271010387157695, 29.70081300813008, 37.69423791821561, 37.26059979317477, 35.30574098798398, 32.18236173393124, 30.947589098532493, 36.46906187624751, 36.20271800679502, 35.42168674698795, 24.9, 27.20416124837451, 33.41676646706587, 35.62522045855379, 33.26258205689278, 42.55678059536935, 38.80923994038748, 27.21664464993395, 31.33783783783784, 30.781710914454276, 33.02304147465438, 35.87307692307692, 24.82110682110682, 29.663522012578618, 33.037441497659906, 27.54982817869416, 24.35777126099707, 34.333767926988266, 29.391975308641975, 33.446175637393765, 32.57551669316375, 30.35531914893617, 26.018979833926455, 26.13929784824462, 31.45762711864407, 35.73848684210526, 33.83765347885402, 35.57567380787837, 33.88341968911917, 28.02880658436214, 30.97911227154047, 40.07978241160471, 44.78774617067834, 36.847983453981385, 33.518181818181816, 35.46263736263736, 31.249773755656108, 33.06426332288401, 55.491379310344826, 38.187732342007436, 27.64332247557003, 117.09826589595376, 86.1]
Plot Mean Area of Nuclei and Mean Intensity
# plot figure
fig = plt.figure(figsize = (16, 5))
gs = GridSpec(nrows = 1, ncols = 2)
ax0 = fig.add_subplot(gs[0, 0])
plt.hist(filledAreaNuclei, bins=30)
ax0 = plt.gca()
ax0.set_title('Mean Area Distribution of Nuclei')
ax0.set_xlabel('mean area / px')
ax0.set_ylabel('number of nuclei')
ax1 = fig.add_subplot(gs[0, 1])
plt.hist(meanIntensityNuclei, bins=30)
ax1 = plt.gca()
ax1.set_title('Mean Intensity Distribution of Nuclei')
ax1.set_xlabel('mean intensity / au')
ax1.set_ylabel('number of nuclei')
Text(0, 0.5, 'number of nuclei')
Goal: Segmentation (first approach) using the Voronoi tesselation.
Steps:
- Compute ellipse-centers on nuclei
- Apply Voronoi tesselation using the ellipse-centers as seeds
masterarray, myImageInclCenters = Groundwork.ellipseFitCenterPos(myMaskContoursAll.astype(np.uint8), myImageRThresholdedRSO)
# print(masterarray)
import sys
import numpy as np
from scipy.spatial import Voronoi, voronoi_plot_2d
from matplotlib import pyplot as plt
from skimage import io, filters
from skimage.feature import peak_local_max
fileName = "02_01_myImageRGB.tif"
myImage = cv2.imread('./Data/' + fileName)
fig = plt.figure(figsize = (15, 10))
ax = fig.add_subplot(111)
ax.imshow(ndimage.rotate(myImageInclCenters, 0))
cv2.circle(myImage, (int(masterarray[3, 0]), int(masterarray[3, 1])), 5, (255, 0, 0), -1)
vor = Voronoi(masterarray)
ax.set_xticks([])
ax.set_yticks([])
plt.show()
plt.savefig('./../ImageExport/Center_of_Masses.png')
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_02_04_Center_of_Masses.png', bbox_inches='tight')
plt.show()
<Figure size 640x480 with 0 Axes>
# sources
# - https://stackoverflow.com/questions/20515554/colorize-voronoi-diagram
# - https://stackoverflow.com/questions/52184271/image-and-voronoi-diagram-on-the-same-figure
# import modules
import matplotlib.pyplot as plt
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
import numpy as np
from scipy.spatial import Voronoi
# compute Voronoi tesselation
vor = Voronoi(masterarray)
# plot
regions, vertices = Groundwork.voronoi_finite_polygons_2d(vor)
fig = plt.figure(figsize=(15, 10))
ax = fig.add_subplot(111)
for region in regions:
polygon = vertices[region]
plt.fill(*zip(*polygon), alpha=0.4)
plt.plot(masterarray[:,0], masterarray[:,1], 'ko')
ax.set_xticks([])
ax.set_yticks([])
plt.imshow(ndimage.rotate(myImageInclCenters, 0))
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/01_02_04_Voronoi_Tesselation.png', bbox_inches='tight')
plt.show()
The results on the given dataset are ok but not remarkable. Still, this algorithm is very robust and one can work with this segmentation: Each of the ellipse centers serves as a seed and the entire set is radially expanded. The points of contact with the neighboring areas limit the evaluation area for each cell. The real strength of the approach is when it comes to dense (i.e. confluent) cell or tissue recordings, where there is hardly any background signal.
Goal:
Analyse a high content microsopic array of fluorescence images in an automated manner. The here used sample is a brain slide with R, G and B labelling. B-Channel encodes the cellular nuclei, which shall be counted for each (artificial) microscopic tile. Each tile simulates the field of view (FOV) of the microscopic setup. R-Channel encodes the GFAP-signal, GFAP is organized in filaments. The filament length shall be measured and quantified computing the average for each FOV. Cell count of the B- and GFAP's average filament length of the R-channel shall be visualized using heatmaps.
Goal:
To determine an appropriate binary mask for nuclei and then perform automated nuclei segmentation.
Step 1: Inspect first excerpt of the brain-slice dataset
# data source
# - http://cellimagelibrary.org/images/CCDB_48367
# Angela Cone (2009) CCDB:48367, NA, None, astrocyte. CIL. Dataset. https://doi.org/doi:10.7295/W9CCDB48367
# data are published under Attribution 3.0 Unported (CC BY 3.0), https://creativecommons.org/licenses/by/3.0/
# rat hippocampus
# red Channel: GFAP
# green Channel: Pannexin 1
# blue Channel: DAPI
# import modules
import cv2
from matplotlib import pyplot as plt
import numpy as np
from scipy.spatial import Voronoi, voronoi_plot_2d
from skimage import io, filters
from skimage.feature import peak_local_max
import sys
# initialize and define variables and parameter
fileName = "48367_excerpt_01.tif"
myImage = cv2.imread("./DataTissue/" + fileName)
# plot figure
fig = plt.figure(figsize = (10, 7))
ax = fig.add_subplot(111)
titleString = "Excerpt of Whole Mouse Brain Sections\n"
plt.title(titleString, fontweight = "bold", fontsize = 14, y = 1.00)
plt.suptitle("DAPI (Blue Channel) / File: " + fileName, fontsize = 10, y = 0.909)
ax.imshow(myImage[:,:,0])
ax.set_xticks([])
ax.set_yticks([])
plt.show()
Step 2: Try edge detection by applying "canny filtering".
Step 3: Try adaptive intensity thresholding. Then combine the results com canny filtering and adaptive thresholding. A dilate-method is applied in order to the gaps, which is necessary to make the binary_fill_holes-function work properly.
Step 4: Elaborate "negative thresholding" in order to extract the nuclei gaps of highly dense regions.
Step 5: Set "background-ones" (step 4) to zero within the binary mask obtained in step 3. Apply split algorithm on the nuclei image.
#####################################
## Step 2 ##
#####################################
# sources
# - https://medium.com/swlh/image-processing-with-python-morphological-operations-26b7006c0359
# import modules
import cv2
from matplotlib import cm
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import make_axes_locatable
from napari_segment_blobs_and_things_with_membranes import threshold_otsu, split_touching_objects
import numpy as np
import pyclesperanto_prototype as cle
from scipy import ndimage
from skimage.color import rgb2gray
from skimage.draw import disk
from skimage.io import imread, imshow
from skimage.morphology import *
# initialize and define variables and parameter
fileName = "48367_excerpt_02.tif"
myImage = cv2.imread("./DataTissue/" + fileName)
myThresholdValue = 120
t_lower = 50 # Lower Threshold
t_upper = 150 # Upper threshold
# compute results
input_image2 = myImage
input_image2[myImage < 50] = 0
edge = cv2.Canny(myImage[:,:,0], t_lower, t_upper)
myImageCannyThresholded = edge
#####################################
## Step 3 ##
#####################################
# sources
# - https://www.youtube.com/watch?v=7u83l_HqamI
# initialize and define variables and parameter
myImage = cv2.imread("./DataTissue/" + fileName)
myImageCopy = cv2.imread("./DataTissue/" + fileName)
myImageCopyThreshold = myImageCopy[:, :, 0]
# compute results
myImageCopyThreshold[myImage[:,:,0] < 50] = 0
myImageAdaptiveThreshold = cv2.adaptiveThreshold(myImageCopyThreshold, 255, cv2.ADAPTIVE_THRESH_MEAN_C, cv2.THRESH_BINARY_INV, 21, 5)
myImageAdaptiveThresholdFilledGaps = ndimage.binary_fill_holes(myImageAdaptiveThreshold).astype(int)
myImageThresholdCombined = np.add(myImageAdaptiveThreshold / 2, myImageCannyThresholded)
# myImageThresholdCombined = myImageAdaptiveThreshold
myImageThresholdUpperOne = np.where(myImageThresholdCombined > 155, 1, 0)
kernel = np.ones((3, 3), np.uint8)
myImageThresholdDilated = cv2.dilate(myImageThresholdUpperOne.astype(np.uint8), None, iterations=1)
myImageThresholdFilledGaps = ndimage.binary_fill_holes(myImageThresholdDilated).astype(int)
#####################################
## Step 4 ##
#####################################
# compute results
myImageThresholdedNeg = np.zeros((myImage.shape[0],myImage.shape[1]))
myImageThresholdedNeg[myImage[:, :, 0] < 20] = 1
# initialize and define variables and parameter
myImageThresholdFilledGaps[myImageThresholdedNeg == 1] = 0
kernel = np.ones((1, 1), np.uint8)
# compute results
myImageTresholdEroded = cv2.erode(myImageThresholdFilledGaps.astype(np.uint8), kernel, iterations=1)
myImageTresholdSplittedObjects = split_touching_objects(myImageTresholdEroded)
myImageSplittedObjects = np.multiply(myImageCopy[:,:,0], myImageTresholdSplittedObjects)
# plot figures
fig, axes = plt.subplots(ncols = 3, nrows = 2, figsize = (17, 11), sharex = True, sharey = True)
ax = axes.ravel()
ii = 0
ax[ii].imshow(myImage[:,:,0], cmap = cm.coolwarm)
ax[ii].set_title('Step 01: Excerpt of Whole Mouse Brain Sections', fontweight = "bold", fontsize = 10)
ax[ii].set_xticks([])
ax[ii].set_yticks([])
ii += 1
ax[ii].imshow(myImageCannyThresholded, cmap = cm.coolwarm)
ax[ii].set_title('Step 02: Binary Mask Canny Detection', fontweight = "bold", fontsize = 10)
ax[ii].set_xticks([])
ax[ii].set_yticks([])
ii += 1
ax[ii].imshow(myImageAdaptiveThresholdFilledGaps, cmap = cm.coolwarm)
ax[ii].set_title('Step 03.1: Binary Mask Intensity Thresholding', fontweight = "bold", fontsize = 10)
ax[ii].set_xticks([])
ax[ii].set_yticks([])
ii += 1
ax[ii].imshow(myImageThresholdFilledGaps, cmap = cm.coolwarm)
ax[ii].set_title('Step 03.2: Combined Mask Intensity \& Canny', fontweight = "bold", fontsize = 10)
ax[ii].set_xticks([])
ax[ii].set_yticks([])
ii += 1
ax[ii].imshow(myImageThresholdedNeg, cmap = cm.coolwarm)
ax[ii].set_title('Step 04: Binary Background Mask', fontweight = "bold", fontsize = 10)
ax[ii].set_xticks([])
ax[ii].set_yticks([])
ii += 1
ax[ii].imshow(myImageSplittedObjects, cmap = cm.coolwarm)
ax[ii].set_title("Step 05: Objected-Splitted Mask", fontweight = "bold", fontsize = 10)
ax[ii].set_xticks([])
ax[ii].set_yticks([])
[]
Simple thresholding or canny detection fails: Thresholding absorbes the intercellular gaps in high density regions, canny detection causes problems when trying to close structures in order to use the binary-fill-function => A combination of both and the background mask works sufficiently well.
Step 6: Apply Voronoi-Otsu-cell-segmentation and show results.
# sources
# - https://datagy.io/python-list-find-all-index/
# import modules
import random
import warnings
warnings.filterwarnings('ignore')
# define extra function
def find_indices(list_to_check, item_to_find):
indices = []
for idx, value in enumerate(list_to_check):
if value == item_to_find:
indices.append(idx)
return indices
# compute results
cleInput = cle.push(myImageSplittedObjects)
sigma_spot_detection = 8
sigma_outline = 1
segmented = cle.voronoi_otsu_labeling(cleInput, spot_sigma = sigma_spot_detection, outline_sigma = sigma_outline)
cellcount = 0
segmented_array = cle.pull(segmented)
maskNucleiAll = np.zeros(segmented_array.shape, np.uint8)
randValue = random.sample(range(np.max(segmented_array)), np.max(segmented_array))
for ii in range(1, np.max(segmented_array)): # 200):
myMaskZeros = np.zeros(segmented_array.shape)
myMaskZeros[segmented_array == ii] = 1
areasize = np.sum(np.sum(myMaskZeros))
if areasize > 500:
myMaskZeros3 = ndimage.binary_fill_holes(myMaskZeros)
maskNucleiTmp = np.zeros(segmented_array.shape, np.uint8)
maskNucleiTmp[myMaskZeros3 == 1] = randValue[ii]
maskNucleiAll = maskNucleiAll + maskNucleiTmp
cellcount += 1
maskNucleiAll[segmented_array == 0] = 0
# plot figure
fig, ax = plt.subplots(figsize = (10, 7))
im = ax.imshow(maskNucleiAll)
plt.title("Voronoi-Otsu-cell-segmentation", fontweight = "bold", fontsize = 14, y = 1.00)
ax.set_xticks([])
ax.set_yticks([])
plt.show()
# print(type(segmented))
# print(cellcount)
Goal:
To determine filament length of GFAP Signal on single image
Step 1: Threshold GFAP signal
Step 2: Label filaments with unique number
Step 3: Eliminate all GFAP fragments in the signal, which are below a certain size. In this way, background signal can be excluded.
# sources
# - https://scikit-image.org/docs/stable/auto_examples/edges/plot_skeleton.html
# - https://www.youtube.com/watch?v=7u83l_HqamI
# import modules
import cv2
import numpy as np
import matplotlib.pyplot as plt
from skimage import data
from skimage.color import rgb2gray
from skimage.draw import disk
from skimage.io import imread, imshow
from skimage.morphology import skeletonize
from skimage.morphology import *
from skimage.util import invert
import os.path
# initialize and define variables and parameter
fileName = "48367_excerpt_02.tif"
myImage = cv2.imread("./DataTissue/" + fileName)
myImageThresholded = np.where(myImage[:, :, 2] > 35, 1, 0)
# compute results
skeletonGFAP = skeletonize(myImageThresholded)
print(np.max(skeletonGFAP))
print(type(skeletonGFAP))
skeletonCorrected = np.zeros(skeletonGFAP.shape)
skeletonCorrected[skeletonGFAP == True] = 1
# # contours, hierarchy = cv2.findContours(skeleton.astype(np.uint8), 1, 2)
# # myImageZeros = np.zeros(skeletonCorrected.shape)
ret, labelsSkeletonGFAP = cv2.connectedComponents(skeletonCorrected.astype(np.uint8))
labelsSkeletonGFAPFilteredAllExcerpt = np.zeros(labelsSkeletonGFAP.shape)
computeData = True
checkFiles = [os.path.exists('./../labelsSkeletonGFAPFilteredAllExcerpt.npy') == True]
if all(checkFiles):
labelsSkeletonGFAPFilteredAllExcerpt = np.load('./../labelsSkeletonGFAPFilteredAllExcerpt.npy', allow_pickle = True)
computeData = False
if computeData == True:
for ii in range(1, ret):
labelTmp = np.zeros(labelsSkeletonGFAP.shape)
labelTmp[labelsSkeletonGFAP == ii] = 1
if np.sum(np.sum(labelTmp)) > 10:
labelsSkeletonGFAPFilteredAllExcerpt = labelsSkeletonGFAPFilteredAllExcerpt + labelTmp
format_float = "{:.2f}".format(ii / ret * 100)
print("Processing ", format_float, "% done", end = "\r")
np.save('./../labelsSkeletonGFAPFilteredAllExcerpt.npy', np.array(labelsSkeletonGFAPFilteredAllExcerpt, dtype = object), allow_pickle = True)
# plot figure
fig, axes = plt.subplots(nrows = 2, ncols = 2, figsize = (12, 10), sharex = True, sharey = True)
ax = axes.ravel()
ax[0].imshow(myImage[:, :, 2], cmap = plt.cm.gray)
ax[0].axis('off')
ax[0].set_title('Excerpt of Whole Mouse Brain Sections: \n GFAP (Red Channel)', fontweight = "bold", fontsize = 10)
ax[1].imshow(skeletonGFAP, cmap = plt.cm.gray)
ax[1].axis('off')
ax[1].set_title('GFAP Skeleton \n', fontweight = "bold", fontsize = 10)
ax[2].imshow(labelsSkeletonGFAP, cmap = plt.cm.gray)
ax[2].axis('off')
ax[2].set_title('Labeled GFAP Skeleton (Red Channel)', fontweight = "bold", fontsize = 10)
ax[3].imshow(labelsSkeletonGFAPFilteredAllExcerpt.astype(np.uint8), cmap = plt.cm.gray)
ax[3].axis('off')
ax[3].set_title('Size-Filtered GFAP Skeleton (Red Channel)', fontweight = "bold", fontsize = 10)
fig.tight_layout()
plt.show()
True <class 'numpy.ndarray'>
Goal: To show segmentation of the blue- (nuclei) and the red- (GFAP) channel at a defined position of the microscopic array
# sources
# - https://scikit-image.org/docs/stable/auto_examples/edges/plot_skeleton.html
# - https://www.youtube.com/watch?v=7u83l_HqamI
# data source
# - http://cellimagelibrary.org/images/CCDB_48367
# Angela Cone (2009) CCDB:48367, NA, None, astrocyte. CIL. Dataset. https://doi.org/doi:10.7295/W9CCDB48367
# data are published under Attribution 3.0 Unported (CC BY 3.0), https://creativecommons.org/licenses/by/3.0/
# rat hippocampus
# import modules
import cv2
import math
from matplotlib import cm
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import make_axes_locatable
from napari_segment_blobs_and_things_with_membranes import threshold_otsu, split_touching_objects
import numpy as np
import pyclesperanto_prototype as cle
import random
from scipy import ndimage
from skimage.color import rgb2gray
from skimage.draw import disk
from skimage.io import imread, imshow
from skimage.morphology import *
import warnings
warnings.filterwarnings('ignore')
# initialize and define variables and parameter
myImage = cv2.imread("./../48367_all.tif")
stepsizeX = 940
stepsizeY = 920
intensity_heatmap = np.zeros((int(math.floor(myImage.shape[0] / stepsizeY)), int(math.floor(myImage.shape[1] / stepsizeX))))
intensity_heatmap2 = np.zeros((int(math.floor(myImage.shape[0] / stepsizeY)), int(math.floor(myImage.shape[1] / stepsizeX))))
jj = 5 # y
ii = 8 # x
# compute results
myImageTmp = myImage[jj * stepsizeY: (jj + 1) * stepsizeY, ii * stepsizeX: (ii + 1) * stepsizeX, :]
myThresholdValue = 125
tLower = 50 # Lower Threshold
tUpper = 150 # Upper threshold
input_image2 = myImageTmp
input_image2[myImageTmp < 50] = 0
edge = cv2.Canny(myImageTmp[:,:,0], tLower, tUpper)
myImageThresholded = edge
myImageCopy= np.copy(myImageTmp)
myImageCopyThreshold = myImageCopy[:, :, 0]
myImageCopyThreshold[myImageCopy[:, :, 0] < 50] = 0
myImageAdaptiveThreshold = cv2.adaptiveThreshold(myImageCopyThreshold, 255, cv2.ADAPTIVE_THRESH_MEAN_C, cv2.THRESH_BINARY_INV, 21, 5)
myImageThresholdCombined = np.add(myImageAdaptiveThreshold / 2, myImageThresholded)
myImageThresholdUpperOne = np.where(myImageThresholdCombined > myThresholdValue, 1, 0)
kernel = np.ones((2, 2), np.uint8)
myImageThresholdDilated = cv2.dilate(myImageThresholdUpperOne.astype(np.uint8), kernel, iterations = 1)
myImageThresholdFilledGaps = ndimage.binary_fill_holes(myImageThresholdDilated).astype(int)
myImageThresholdedNeg = np.zeros((myImageTmp.shape[0],myImageTmp.shape[1]))
myImageThresholdedNeg[myImageTmp[:, :, 0] < 20] = 1
myImageThresholdFilledGaps[myImageThresholdedNeg == 1] = 0
kernel = np.ones((1, 1), np.uint8)
myImageThresholdEroded = cv2.erode(myImageThresholdFilledGaps.astype(np.uint8), kernel, iterations=1)
myImageThresholdSplittedObjects = split_touching_objects(myImageThresholdEroded)
myImageSplittedObjects = np.multiply(myImageCopy[:,:,0], myImageThresholdSplittedObjects)
cleInput = cle.push(myImageSplittedObjects)
sigmaSpotDetection = 8
sigmaOutline = 1
segmented = cle.voronoi_otsu_labeling(cleInput, spot_sigma = sigmaSpotDetection, outline_sigma = sigmaOutline)
cellcount = 0
segmentedArray = cle.pull(segmented)
maskNucleiAll = np.zeros(segmentedArray.shape, np.uint8)
randValue = random.sample(range(np.max(segmentedArray)), np.max(segmentedArray))
for kk in range(1, np.max(segmented_array)):#200):
myMaskZeros = np.zeros(segmentedArray.shape)
myMaskZeros[segmentedArray == kk] = 1
areasize = np.sum(np.sum(myMaskZeros))
if areasize > 500:
maskNucleiFilledHolesTmp = ndimage.binary_fill_holes(myMaskZeros)
maskNucleiTmp = np.zeros(segmentedArray.shape, np.uint8)
maskNucleiTmp[maskNucleiFilledHolesTmp == 1] = randValue[kk]
maskNucleiAll = maskNucleiAll + maskNucleiTmp
cellcount += 1
maskNucleiAll[segmentedArray == 0] = 0
intensity_heatmap[jj, ii] = cellcount
myImageThresholded = np.where(myImageTmp[:,:,2] > 35, 1, 0)
skeleton = skeletonize(myImageThresholded)
skeletonCorrected = np.zeros(skeleton.shape)
skeletonCorrected[skeleton == True] = 1
ret, labelsSkeletonGFAP = cv2.connectedComponents(skeletonCorrected.astype(np.uint8))
labelsSkeletonGFAPFilteredAll = np.zeros(labelsSkeletonGFAP.shape)
for ll in range(1, ret):
labelTmp = np.zeros(labelsSkeletonGFAP.shape)
labelTmp[labelsSkeletonGFAP == ll] = 1
if np.sum(np.sum(labelTmp)) > 10:
labelsSkeletonGFAPFilteredAll = labelsSkeletonGFAPFilteredAll + labelTmp
intensity_heatmap2[jj, ii] = np.sum(np.sum(labelsSkeletonGFAPFilteredAll))
myImageCopy= np.copy(myImageTmp)
myImageCopy2 = np.copy(myImage)
# mark current position in myImage-representation
myImageCopy2= np.copy(myImage)
myImageCopy2[jj * stepsizeY: (jj + 1) * stepsizeY, ii * stepsizeX: (ii + 1) * stepsizeX, 0] = 255
myImageCopy3 = np.copy(myImage)
# print results
dashline_length = 41
print("-" * dashline_length, end = '\n')
print(" " * 15, "RESULTS", end = '\n')
print("-" * dashline_length, end = '\n')
print(" " * 11, "array attributes", end = '\n')
print("size: \t\t\t", myImage.shape)
print("# tiles x-direction: \t", int(math.floor(myImage.shape[0] / stepsizeX)))
print("# tiles y-direction: \t", int(math.floor(myImage.shape[1] / stepsizeY)))
print("-" * dashline_length, end = '\n')
print(" " * 10, "count for analysis", end = '\n')
print("cellcount: \t\t", cellcount)
print("filament length: \t", np.sum(np.sum(labelsSkeletonGFAPFilteredAll)))
print("-" * dashline_length, end = '\n')
# plot figure
fig, ax = plt.subplots(figsize = (15, 10))
im = ax.imshow(myImageCopy2)
plt.title("Entire Micrograph Tile Array", fontweight = "bold", fontsize = 16, y = 1.03)
plt.suptitle("(Current Position is Marked by Red Rectangle)", fontsize = 10, y = 0.87)
ax.set_xticks([])
ax.set_yticks([])
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/02_03_Micrograph_Tile_Array01.png', bbox_inches='tight')
plt.show()
fig, axes = plt.subplots(nrows = 2, ncols = 2, figsize = (10, 10), sharex = True, sharey = True)
ax = axes.ravel()
currentFOV = myImageCopy3[jj * stepsizeY: (jj + 1) * stepsizeY, ii * stepsizeX: (ii + 1) * stepsizeX,:]
ax[0].imshow(currentFOV, cmap = plt.cm.gray)
ax[0].axis('off')
ax[0].set_title('Current FOV', fontweight = "bold", fontsize = 10)
ax[1].imshow(currentFOV[:,:,0], cmap = plt.cm.gray)
ax[1].axis('off')
ax[1].set_title('Current FOV (Blue Channel)', fontweight = "bold", fontsize = 10)
ax[2].imshow(maskNucleiAll, cmap = plt.cm.gray)
ax[2].axis('off')
ax[2].set_title('Nuclei Labeled', fontweight = "bold", fontsize = 10)
ax[3].imshow(labelsSkeletonGFAPFilteredAll, cmap = plt.cm.gray)
ax[3].axis('off')
ax[3].set_title('GFAP Skeleton', fontweight = "bold", fontsize = 10)
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/02_03_Micrograph_Tile_Array02.png', bbox_inches='tight')
plt.show()
-----------------------------------------
RESULTS
-----------------------------------------
array attributes
size: (9718, 15910, 3)
# tiles x-direction: 10
# tiles y-direction: 17
-----------------------------------------
count for analysis
cellcount: 160
filament length: 13294.0
-----------------------------------------
Goal: To compute nuclei and GFAP-filament integrated intensity on each tile of the microscopic array. Visualize results of this co-localization experiment in a heatmap.
# sources
# - https://scikit-image.org/docs/stable/auto_examples/edges/plot_skeleton.html
# - https://www.youtube.com/watch?v=7u83l_HqamI
# import modules
import cv2
import math
import matplotlib.pyplot as plt
import numpy as np
from skimage.color import rgb2gray
from skimage.draw import disk
from skimage.io import imread, imshow
from skimage.morphology import *
# initialize and define variables and parameter
myImage = cv2.imread("./../48367_all.tif")
stepsizeX = 300
stepsizeY = 300
intensity_heatmap = np.zeros((int(math.floor(myImage.shape[0] / stepsizeY)), int(math.floor(myImage.shape[1] / stepsizeX))))
intensity_heatmap2 = np.zeros((int(math.floor(myImage.shape[0] / stepsizeY)), int(math.floor(myImage.shape[1] / stepsizeX))))
# compute results
for jj in range(int(math.floor(myImage.shape[0] / stepsizeY)) - 1):
for ii in range(int(math.floor(myImage.shape[1] / stepsizeX)) - 1):
im_curr = myImage[jj * stepsizeY: (jj + 1) * stepsizeY, ii * stepsizeX: (ii + 1) * stepsizeX, :]
intensity_heatmap[jj, ii] = np.sum(np.sum(im_curr[:, :, 0]))
# if ii==3 and jj==3:
# fig, ax = plt.subplots(figsize = (15, 10))
# im = ax.imshow(im_curr)
# plt.title("Distance Transform", fontweight="bold", fontsize = 16, y = 1.05)
# ax.set_xticks([])
# ax.set_yticks([])
# plt.show()
intensity_heatmap2[jj, ii] = np.sum(np.sum(im_curr[:, :, 2]))
# plot figure
fig, axes = plt.subplots(nrows = 1, ncols = 2, figsize = (15, 10), sharex = True, sharey = True)
ax = axes.ravel()
ax[0].imshow(intensity_heatmap, cmap = plt.cm.gray)
ax[0].axis('off')
ax[0].set_title('Nuclei Integrated Intensity', fontweight = "bold", fontsize = 14)
ax[1].imshow(intensity_heatmap2, cmap = plt.cm.gray)
ax[1].axis('off')
ax[1].set_title('GFAP Integrated Intensity', fontweight = "bold", fontsize = 14)
fig.tight_layout()
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/02_04_Micrograph_Tile_Array03.png', bbox_inches='tight')
plt.show()
Goal: To compute nuclei count and GFAP-filament length on each tile of the microscopic array. Visualize results in a heatmap.
# sources
# - https://scikit-image.org/docs/stable/auto_examples/edges/plot_skeleton.html
# - https://www.youtube.com/watch?v=7u83l_HqamI
# - https://stackoverflow.com/questions/49367144/modify-matplotlib-colormap
# import modules
import cv2
import math
from matplotlib import cm
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
import matplotlib as mpl
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
from mpl_toolkits.axes_grid1 import make_axes_locatable
from napari_segment_blobs_and_things_with_membranes import threshold_otsu, split_touching_objects
from numpy import loadtxt
import numpy as np
from numpy import savetxt
import os.path
import pyclesperanto_prototype as cle
import random
from scipy import ndimage
from skimage.color import rgb2gray
from skimage.draw import disk
from skimage.io import imread, imshow
from skimage.morphology import *
import warnings
warnings.filterwarnings('ignore')
# initialize and define variables and parameter
nucleiMaskEntireDataset = []
nucleiMaskEntireDatasetAllFields = []
GFAPSkeletonEntireDataset = []
stepsizeX = 940
stepsizeY = 920
myImage = cv2.imread("./../48367_all.tif")
intensity_heatmap = np.zeros((int(math.floor(myImage.shape[0] / stepsizeY)), int(math.floor(myImage.shape[1] / stepsizeX))))
intensity_heatmap2 = np.zeros((int(math.floor(myImage.shape[0] / stepsizeY)), int(math.floor(myImage.shape[1] / stepsizeX))))
# print(myImage.shape)
# print(int(math.floor(myImage.shape[0] / stepsizeX)))
# print(int(math.floor(myImage.shape[1] / stepsizeY)))
computeData = True
checkFiles = [os.path.exists('./../DATA/intensity_heatmap.csv') == True,
os.path.exists('./../DATA/intensity_heatmap2.csv') == True,
os.path.exists('./../DATA/nucleiMaskEntireDataset.npy') == True,
os.path.exists('./../DATA/nucleiMaskEntireDatasetAllFields.npy') == True,
os.path.exists('./../DATA/GFAPSkeletonEntireDataset.npy') == True]
if all(checkFiles):
intensity_heatmap = loadtxt('./../DATA/intensity_heatmap.csv', delimiter = ',')
intensity_heatmap2 = loadtxt('./../DATA/intensity_heatmap2.csv', delimiter = ',')
nucleiMaskEntireDataset = np.load('./../DATA/nucleiMaskEntireDataset.npy', allow_pickle = True)
GFAPSkeletonEntireDataset = np.load('./../DATA/GFAPSkeletonEntireDataset.npy', allow_pickle = True)
nucleiMaskEntireDatasetAllFields = np.load('./../DATA/nucleiMaskEntireDatasetAllFields.npy', allow_pickle = True)
computeData = False
if computeData == True:
cellcountAll = 1
# compute results
for jj in range(int(math.floor(myImage.shape[0] / stepsizeY))):
for ii in range(int(math.floor(myImage.shape[1] / stepsizeX))):
myImageTmp = myImage[jj * stepsizeY: (jj + 1) * stepsizeY, ii * stepsizeX: (ii + 1) * stepsizeX, :]
myThresholdValue = 125
t_lower = 50 # Lower Threshold
t_upper = 150 # Upper threshold
input_image2 = myImageTmp
input_image2[myImageTmp < 50] = 0
edge = cv2.Canny(myImageTmp[:,:,0], t_lower, t_upper)
myImageThresholded = edge
myImageCopy= np.copy(myImageTmp)
myImageCopyTreshold = myImageCopy[:, :, 0]
myImageCopyTreshold[myImageCopy[:, :, 0] < 50] = 0
myImageAdaptiveTreshold = cv2.adaptiveThreshold(myImageCopyTreshold, 255, cv2.ADAPTIVE_THRESH_MEAN_C, cv2.THRESH_BINARY_INV, 21, 5)
myImageTresholdCombined = np.add(myImageAdaptiveTreshold / 2, myImageThresholded)
myImageTresholdUpperOne = np.where(myImageTresholdCombined > myThresholdValue, 1, 0)
kernel = np.ones((2, 2), np.uint8)
myImageTresholdDilated = cv2.dilate(myImageTresholdUpperOne.astype(np.uint8), kernel, iterations = 1)
myImageTresholdFilledGaps = ndimage.binary_fill_holes(myImageTresholdDilated).astype(int)
myImageThresholdedNeg = np.zeros((myImageTmp.shape[0],myImageTmp.shape[1]))
myImageThresholdedNeg[myImageTmp[:, :, 0] < 20] = 1
myImageTresholdFilledGaps[myImageThresholdedNeg == 1] = 0
kernel = np.ones((1, 1), np.uint8)
myImageTresholdEroded = cv2.erode(myImageTresholdFilledGaps.astype(np.uint8), kernel, iterations=1)
myImageTresholdSplittedObjects = split_touching_objects(myImageTresholdEroded)
myImageSplittedObjects = np.multiply(myImageCopy[:,:,0], myImageTresholdSplittedObjects)
input_gpu = cle.push(myImageSplittedObjects)
sigma_spot_detection = 8
sigma_outline = 1
segmented = cle.voronoi_otsu_labeling(input_gpu, spot_sigma = sigma_spot_detection, outline_sigma = sigma_outline)
cellcount = 0
segmented_array = cle.pull(segmented)
maskNucleiAll = np.zeros(segmented_array.shape, np.uint8)
maskNucleiAllFields = np.zeros(segmented_array.shape, np.uint8)
randValue = random.sample(range(np.max(segmented_array)), np.max(segmented_array))
for kk in range(1, np.max(segmented_array)):#200):
myMaskZeros = np.zeros(segmented_array.shape)
myMaskZeros[segmented_array == kk] = 1
areasize = np.sum(np.sum(myMaskZeros))
if areasize > 500:
maskNucleiFilledHolesTmp = ndimage.binary_fill_holes(myMaskZeros)
maskNucleiTmp = np.zeros(segmented_array.shape, np.uint8)
maskNucleiTmpAllFields = np.zeros(segmented_array.shape)
maskNucleiTmp[maskNucleiFilledHolesTmp == 1] = randValue[kk]
maskNucleiAll = maskNucleiAll + maskNucleiTmp
maskNucleiTmpAllFields[maskNucleiFilledHolesTmp == 1] = cellcountAll
x1_rounded, y1_rounded = Groundwork.centerOfMass(maskNucleiTmpAllFields)
maskNucleiAllFields = maskNucleiAllFields + maskNucleiTmpAllFields
# print(np.max(maskNucleiAll))
# print("maskNucleiAllFields: ",np.max(maskNucleiAllFields))
cellcount += 1
cellcountAll += 1
maskNucleiAll[segmented_array == 0] = 0
maskNucleiAllFields[segmented_array == 0] = 0
nucleiMaskEntireDataset.append(maskNucleiAll)
nucleiMaskEntireDatasetAllFields.append(maskNucleiAllFields)
intensity_heatmap[jj, ii] = cellcount
myImageThresholded = np.where(myImageTmp[:, :, 2] > 35, 1, 0)
skeleton = skeletonize(myImageThresholded)
skeletonCorrected = np.zeros(skeleton.shape)
skeletonCorrected[skeleton == True] = 1
ret, labelsSkeletonGFAP = cv2.connectedComponents(skeletonCorrected.astype(np.uint8))
labelsSkeletonGFAPFilteredAll = np.zeros(labelsSkeletonGFAP.shape)
for ll in range(1, ret):
labelTmp = np.zeros(labelsSkeletonGFAP.shape)
labelTmp[labelsSkeletonGFAP == ll] = 1
if np.sum(np.sum(labelTmp)) > 10:
labelsSkeletonGFAPFilteredAll = labelsSkeletonGFAPFilteredAll + labelTmp
intensity_heatmap2[jj, ii] = np.sum(np.sum(labelsSkeletonGFAPFilteredAll))
GFAPSkeletonEntireDataset.append(labelsSkeletonGFAPFilteredAll)
format_float = "{:.2f}".format((jj + 1) * stepsizeY / (myImage.shape[0]) * 100)
print("Processing ", format_float, "% done", end = "\r")
print(" " * 100, end = "\r")
print("Processing 100% done", end = "\r")
savetxt('./../DATA/intensity_heatmap.csv', intensity_heatmap, delimiter = ',')
savetxt('./../DATA/intensity_heatmap2.csv', intensity_heatmap2, delimiter = ',')
np.save('./../DATA/nucleiMaskEntireDataset.npy', np.array(nucleiMaskEntireDataset, dtype = object), allow_pickle = True)
np.save('./../DATA/GFAPSkeletonEntireDataset.npy', np.array(GFAPSkeletonEntireDataset, dtype = object), allow_pickle = True)
np.save('./../DATA/nucleiMaskEntireDatasetAllFields.npy', np.array(nucleiMaskEntireDatasetAllFields, dtype = object), allow_pickle = True)
upper = mpl.cm.jet(np.arange(256))
lower = np.ones((int(256/256),4))
for i in range(3):
lower[:,i] = np.linspace(1, upper[0,i], lower.shape[0])
cmap = np.vstack(( lower, upper ))
jetWhite = mpl.colors.ListedColormap(cmap, name='myColorMap', N=cmap.shape[0])
# plot figure
fig, axes = plt.subplots(nrows = 1, ncols = 2, figsize = (15, 10), sharex = True, sharey = True)
ax = axes.ravel()
fig0 = ax[0].imshow(intensity_heatmap, cmap = jetWhite) #plt.cm.jet)
fig.colorbar(fig0, ax=ax[0], location='bottom')
ax[0].axis('off')
ax[0].set_title('Nuclei-Count for Each Micrograph Tile', fontweight = "bold", fontsize = 14)
fig1 = ax[1].imshow(intensity_heatmap2, cmap = jetWhite) #plt.cm.jet)
fig.colorbar(fig1, ax=ax[1], location='bottom')
ax[1].axis('off')
ax[1].set_title('Skeleton-Length for Each Micrograph Tile', fontweight = "bold", fontsize = 14)
fig.tight_layout()
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/02_04_Micrograph_Tile_Array04.png', bbox_inches='tight')
plt.show()
Goal: To plot all nuclei masks and GFAP-skeletons of the micrograph tile array. The cellcount of each tile shall be written as a number into the tile image.
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
nucleiMaskEntireDatasetOneImage = np.zeros((myImage.shape[0],myImage.shape[1]))
GFAPSkeletonEntireDatasetOneImage = np.zeros((myImage.shape[0],myImage.shape[1]))
nucleiMaskEntireDatasetOneImageAllFields = np.zeros((myImage.shape[0],myImage.shape[1]))
countTiles = 0
print(nucleiMaskEntireDatasetOneImage.shape)
for jj in range(int(math.floor(myImage.shape[0] / stepsizeY))):
for ii in range(int(math.floor(myImage.shape[1] / stepsizeX))):
nucleiMaskEntireDatasetOneImage[jj * stepsizeY: (jj+1) * (stepsizeY), ii * stepsizeX: (ii + 1) * (stepsizeX)] = nucleiMaskEntireDataset[countTiles]
nucleiMaskEntireDatasetOneImageAllFields[jj * stepsizeY: (jj+1) * (stepsizeY), ii * stepsizeX: (ii + 1) * (stepsizeX)] = nucleiMaskEntireDatasetAllFields[countTiles]
GFAPSkeletonEntireDatasetOneImage[jj * stepsizeY: (jj+1) * (stepsizeY), ii * stepsizeX: (ii + 1) * (stepsizeX)] = GFAPSkeletonEntireDataset[countTiles]
countTiles += 1
for jj in range(int(math.floor(myImage.shape[0] / stepsizeY))):
for ii in range(int(math.floor(myImage.shape[1] / stepsizeX))):
xzo = int(ii * stepsizeX + round(stepsizeX / 2 - 0.05 * stepsizeX))
yzo = int(jj * stepsizeY + round(stepsizeY / 2 - 0.05 * stepsizeY))
cv2.putText(nucleiMaskEntireDatasetOneImage, str(int(intensity_heatmap[jj, ii])), (xzo ,yzo), cv2.FONT_HERSHEY_COMPLEX, 10, (200, 200, 0), 2)
# plot figure
fig, axes = plt.subplots(nrows = 2, ncols = 1, figsize = (15, 10), sharex = True, sharey = True)
ax = axes.ravel()
ax[0].imshow(nucleiMaskEntireDatasetOneImage, cmap = jetWhite) #plt.cm.jet)
ax[0].axis('off')
ax[0].set_title('Nuclei Masks (Entire Microscopic Tile Array)', fontweight = "bold", fontsize = 14)
ax[1].imshow(GFAPSkeletonEntireDatasetOneImage, cmap = plt.cm.binary)
ax[1].axis('off')
ax[1].set_title('GFAP Skeletons (Entire Microscopic Tile Array)', fontweight = "bold", fontsize = 14)
fig.tight_layout()
plt.savefig('./../ImageExport/TissueMasks.png', dpi = 2400)
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/02_03_Micrograph_Tile_Array05.png', bbox_inches='tight', dpi=300)
plt.show()
(9718, 15910)
Remarks: Nuclei on the borders of each tile are potentially counted twice. If one is picky, this error has to be excluded. Typically this is done by artificially shrinking the tiles, e.g. on the right and bottom edge. An even better way could be to trace nuclei on the entire stitched tile dataset as a whole. Due to a lack of computation power, this approach is not considered here. For even larger datasets this approach can become even more tricky. Still, the results shown above are reasonable, since a few cells less have no significant role in statistics.
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
fig, ax = plt.subplots(nrows = 1, ncols = 1, figsize = (10, 7), sharex = True, sharey = True)
# ax = axes.ravel()
fig0 = ax.imshow(nucleiMaskEntireDatasetOneImageAllFields, cmap = jetWhite) #plt.cm.jet)
divider = make_axes_locatable(ax)
cax = divider.append_axes('right', size="2%", pad=0.05)
plt.colorbar(fig0, cax=cax)
ax.axis('off')
ax.set_title('Nuclei Masks (Entire Microscopic Tile Array) with encounting numbers', fontweight = "bold", fontsize = 14)
fig.tight_layout()
# plt.savefig('./../ImageExport/TissueMasks.png', dpi = 2400)
plt.show()
Goal: To extract high density cellular regions such as the hypocampus of brain slices by next-neighbor-counting for each cell of the nuclei mask
Step 1: Determine the center-of-mass (with coordinates x and y) for each cell on the binary mask.
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
computeData = True
checkFiles = [os.path.exists('./../DATA/xc_all.npy') == True,
os.path.exists('./../DATA/yc_all.npy') == True]
if all(checkFiles):
xc_all = np.load('./../DATA/xc_all.npy', allow_pickle = True)
yc_all = np.load('./../DATA/yc_all.npy', allow_pickle = True)
computeData = False
if computeData == True:
xc_all = []
yc_all = []
for ii in range(1, int(np.max(nucleiMaskEntireDatasetOneImageAllFields))):
nucleiAllCenterOfMass = np.zeros(nucleiMaskEntireDatasetOneImageAllFields.shape)
nucleiAllCenterOfMass[nucleiMaskEntireDatasetOneImageAllFields==ii] = 1
x1_rounded, y1_rounded = Groundwork.centerOfMass(nucleiAllCenterOfMass)
xc_all.append(x1_rounded)
yc_all.append(y1_rounded)
print(ii/int(np.max(nucleiMaskEntireDatasetOneImageAllFields)), end="\r")
# sleep 0.02
del nucleiAllCenterOfMass
np.save('./../DATA/xc_all.npy', np.array(xc_all, dtype = object), allow_pickle = True)
np.save('./../DATA/yc_all.npy', np.array(yc_all, dtype = object), allow_pickle = True)
Step 2: Determine all cells which have 10 neighbors within a 100 pixel radius (Euclidean distance).
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
# in pixel
# maxDist = 20
maxRadius = 100
minNeighborCells = 10
highDensityList = []
countNeighborsAll = []
computeData = True
checkFiles = [os.path.exists('./../DATA/countNeighborsAll_maxRadius_'+str(maxRadius)+'_minNeighborCells_'+str(minNeighborCells)+'.npy') == True,
os.path.exists('./../DATA/highDensityList_maxRadius_'+str(maxRadius)+'_minNeighborCells_'+str(minNeighborCells)+'.npy') == True]
if all(checkFiles):
countNeighborsAll = np.load('./../DATA/countNeighborsAll_maxRadius_'+str(maxRadius)+'_minNeighborCells_'+str(minNeighborCells)+'.npy', allow_pickle = True)
highDensityList = np.load('./../DATA/highDensityList_maxRadius_'+str(maxRadius)+'_minNeighborCells_'+str(minNeighborCells)+'.npy', allow_pickle = True)
computeData = False
if computeData == True:
for ii in range(len(yc_all)):
xcurr = xc_all[ii]
ycurr = yc_all[ii]
countNeighbors = 0
for jj in range(len(yc_all)):
if (xcurr-xc_all[jj])**2 + (ycurr-yc_all[jj])**2 <= maxRadius**2:
countNeighbors += 1
countNeighborsAll.append(countNeighbors)
if countNeighbors >= minNeighborCells:
highDensityList.append(1)
else:
highDensityList.append(0)
print(ii/int(np.max(nucleiMaskEntireDatasetOneImageAllFields)), end="\r")
np.save('./../DATA/countNeighborsAll_maxRadius_'+str(maxRadius)+'_minNeighborCells_'+str(minNeighborCells)+'.npy', np.array(countNeighborsAll, dtype = object), allow_pickle = True)
np.save('./../DATA/highDensityList_maxRadius_'+str(maxRadius)+'_minNeighborCells_'+str(minNeighborCells)+'.npy', np.array(highDensityList, dtype = object), allow_pickle = True)
Step 3: Show the two cell subcategories in a scatter plot.
- blue: Low density regions
- red: high density regions fullfilling the "high density criterion" of step 2
import numpy
import matplotlib.pyplot as plt
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
fig, ax = plt.subplots()
for ii in range(len(yc_all)):
if highDensityList[ii] == 1:
ax.scatter(xc_all[ii], yc_all[ii], c='r', s=0.3)
else:
ax.scatter(xc_all[ii], yc_all[ii], c='b', s=0.3)
print(ii/int(np.max(nucleiMaskEntireDatasetOneImageAllFields)), end="\r")
ax.set_xlim(0, 15500)
ax.set_ylim(0, 9000)
plt.gca().invert_yaxis()
plt.tight_layout()
plt.savefig('./../ImageExport/highDensityPlotCells_'+str(maxRadius)+'_minNeighborCells_'+str(minNeighborCells)+'.png', dpi = 600)
plt.show()
0.99976955870491995656
# import modules
import cv2
import numpy as np
import matplotlib.pyplot as plt
from skimage.measure import regionprops
from statistics import mean
from matplotlib.gridspec import GridSpec
binsNumber = 21
# plot figure
fig = plt.figure(figsize = (11, 5))
gs = GridSpec(nrows = 1, ncols = 1)
listXLabel = []
for ii in range(0,binsNumber):
listXLabel.append(str(ii))
ax0 = fig.add_subplot(gs[0, 0])
n, bins, patches = plt.hist(countNeighborsAll, bins=22)
for ii in range(0,binsNumber):
if ii >= minNeighborCells-1:
patches[ii].set_fc('r')
else:
patches[ii].set_fc('b')
ax0 = plt.gca()
min_ylim, max_ylim = plt.ylim()
plt.axvline(minNeighborCells, color='k', linestyle='dashed', linewidth = 1)
plt.text(10.2, 1400, 'Border nuclei of high and low density regions')
ax0.set_title('Next Neighbor Count Distribution Within ' + str(maxRadius) + ' Pixel Radius')
ax0.set_xlabel('next neighbors within ' + str(maxRadius) +' pixel radius')
ax0.set_ylabel('nuclei count')
ax0.set_xticks(np.arange(0.5, binsNumber+0.5, 1), listXLabel)
ax0.set_yticks(np.arange(0, 1600, 100))
ax0.set_xlim(0,binsNumber)
ax0.set_ylim(0,1500)
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/countNeighborsAll.png', bbox_inches='tight')
plt.show()
Goal: Analyse a time series recording of cells with respect to contour / shape and local degree of curvature
Step 1: Thresholding
# sources
# - https://stackabuse.com/opencv-edge-detection-in-python-with-cv2canny/
# - http://www.learningaboutelectronics.com/Articles/How-to-find-the-largest-or-smallest-object-in-an-image-Python-OpenCV.php
# data source
# Rachel Fink, Pat Wadsworth (2011) CIL:35204, Fundulus heteroclitus, deep cell. CIL. Dataset. https://doi.org/doi:10.7295/W9CIL35204
# - http://cellimagelibrary.org/images/35204
# data are published under (CC BY-NC-SA 3.0) , https://creativecommons.org/licenses/by-nc-sa/3.0/
# import modules
import cv2
from matplotlib import cm
from matplotlib.gridspec import GridSpec
import matplotlib.pyplot as plt
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
from mpl_toolkits.axes_grid1 import make_axes_locatable
import numpy as np
from scipy.ndimage import gaussian_filter
from scipy import ndimage
import time
# contour determination
def get_contour_areas(contours):
all_areas= []
for cnt in contours:
area = cv2.contourArea(cnt)
all_areas.append(area)
return all_areas
# initialize and define variables and parameter
filename = "./DataExternalMov/35204.mov"
cap = cv2.VideoCapture(filename)
frames_all = []
start = time.time()
cap = cv2.VideoCapture(filename)
i = 40
success = cap.grab()
ret, image = cap.retrieve()
kernel = np.ones((2, 2), np.uint8)
myImageGray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
# threshold processing
myImageThresholded = np.zeros(myImageGray.shape)
myThresholdValue = 210
myImageThresholded[myImageGray > myThresholdValue] = 1
t_lower = 210
t_upper = 250
aperture_size = 3
cannyImg = cv2.Canny(myImageGray, t_lower, t_upper, apertureSize = aperture_size)
dilation = cv2.dilate(cannyImg, kernel, iterations = 2)
dilation[myImageThresholded == 1] = 255
cannyImgFill = ndimage.binary_fill_holes(dilation).astype(int)
cannyImgRGB = np.zeros(image.shape)
cannyImgRGB[:,:,0] = cannyImgFill * 255
cannyImgRGBConv = np.asarray(cannyImgRGB, np.uint8)
# contour determination
contours, hierarchy = cv2.findContours(cannyImgFill.astype(np.uint8), 1, 1)
sorted_contours = sorted(contours, key=cv2.contourArea, reverse= True)
largest_item = sorted_contours[0]
myMaskContoursAllCurvatureValues = cv2.drawContours(image, largest_item, -1, (255,255,255),1)
contoursxy = np.zeros((len(largest_item), 2))
ll = list(largest_item)
contoursx = []
contoursy = []
for ii in range(len(ll)):
contoursx.append(ll[ii][0][0])
contoursy.append(ll[ii][0][1])
if len(contoursxy) > 1:
contoursxy[:,0] = contoursx
contoursxy[:,1] = contoursy
contourCurvature, contourPixels = Groundwork.calculateContourCurvature(contoursxy)
y = contourCurvature
yhat = gaussian_filter(y, sigma = 10)
for ii in range(len(contourPixels)):
colorValue = 255*yhat[ii]/max(yhat)
if np.isnan(colorValue) == False:
colorPos = int(round(colorValue))
rcolor = int(round(cm.jet(colorPos)[0]*255))
gcolor = int(round(cm.jet(colorPos)[1]*255))
bcolor = int(round(cm.jet(colorPos)[2]*255))
rgbcolor = [rcolor, gcolor, bcolor]
thickness = 1
myMaskContoursAllCurvatureValues[int(contoursxy[ii,1])-thickness:int(contoursxy[ii,1])+thickness,int(contoursxy[ii,0])-thickness:int(contoursxy[ii,0])+thickness,:] = rgbcolor
added_image = cv2.addWeighted(myMaskContoursAllCurvatureValues, 0.95, cannyImgRGBConv, 0.01, 0)
frames_all.append(added_image)
fig = plt.figure(figsize = (7, 5))
gs = GridSpec(nrows = 1, ncols = 1)
ax0 = fig.add_subplot(gs[0, 0])
plt.imshow(added_image)
ax0 = plt.gca()
ax0.set_title('Fundulus heteroclitus embryo during gastrulation', fontweight = "bold", fontsize = 14)
if saveGallery == True:
plt.savefig('./../GALLERY/Analysis_Of_BioImages/03_01_Time_Lapse_Curvature01.png', bbox_inches='tight')
plt.show()
Step 2:
- Load video and process frame by frame
- Threshold with the values obtained from above
- Apply contour-finding on the threshold binary mask
- Colorcode the contour with curvature values using jetmap-colorcoding
- Overlay with micrograph
- Export as video
# sources
# - http://www.learningaboutelectronics.com/Articles/How-to-find-the-largest-or-smallest-object-in-an-image-Python-OpenCV.php
# - https://stackabuse.com/opencv-edge-detection-in-python-with-cv2canny/
# - https://z-uo.medium.com/opencv-automatic-select-big-contour-areas-and-remove-8d79464a06e7
# data source
# Rachel Fink, Pat Wadsworth (2011) CIL:35204, Fundulus heteroclitus, deep cell. CIL. Dataset. https://doi.org/doi:10.7295/W9CIL35204
# - http://cellimagelibrary.org/images/35204
# data are published under (CC BY-NC-SA 3.0) , https://creativecommons.org/licenses/by-nc-sa/3.0/
# import modules
import cv2
import imageio
from matplotlib import cm
import matplotlib.pyplot as plt
import ModulesOwnAndExternalModified.A_Groundwork as Groundwork
from mpl_toolkits.axes_grid1 import make_axes_locatable
import numpy as np
from scipy import ndimage
from scipy.ndimage import gaussian_filter
import time
import warnings
warnings.filterwarnings('ignore')
# contour determination
def get_contour_areas(contours):
all_areas= []
for cnt in contours:
area = cv2.contourArea(cnt)
all_areas.append(area)
return all_areas
# initialize and define variables and parameter
filename = "./DataExternalMov/35204.mov"
cap = cv2.VideoCapture(filename)
frames_all = []
start = time.time()
cap = cv2.VideoCapture(filename)
for i in range(0, 84):
success = cap.grab()
ret, image = cap.retrieve()
kernel = np.ones((2, 2), np.uint8)
myImageGray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
# threshold processing
myImageThresholded = np.zeros(myImageGray.shape)
myThresholdValue = 140
myImageThresholded[myImageGray > myThresholdValue] = 1
t_lower = 210
t_upper = 250
aperture_size = 3
myImageThresholdedDilation = cv2.dilate(myImageThresholded, kernel, iterations = 2)
myImageThresholdedDilationFill = ndimage.binary_fill_holes(myImageThresholdedDilation).astype(int)
myImageThresholdedRGB = np.zeros(image.shape)
myImageThresholdedRGB[:,:,0] = myImageThresholdedDilationFill * 255
myImageThresholdedRGBConv = np.asarray(myImageThresholdedRGB, np.uint8)
# contour determination
contours, hierarchy = cv2.findContours(myImageThresholdedDilationFill.astype(np.uint8), 1, 1)
sorted_contours = sorted(contours, key = cv2.contourArea, reverse = True)
largest_item = sorted_contours[0]
myMaskContoursAllCurvatureValues = cv2.drawContours(image, largest_item, -1, (255,255,255), 1)
contoursxy = np.zeros((len(largest_item), 2))
ll = list(largest_item)
contoursx = []
contoursy = []
for ii in range(len(ll)):
contoursx.append(ll[ii][0][0])
contoursy.append(ll[ii][0][1])
if len(contoursxy) > 1:
contoursxy[:,0] = contoursx
contoursxy[:,1] = contoursy
contourCurvature, contourPixels = Groundwork.calculateContourCurvature(contoursxy)
y = contourCurvature
yhat = gaussian_filter(y, sigma = 10)
for ii in range(len(contourPixels)):
colorValue = 255 * yhat[ii] / max(yhat)
if np.isnan(colorValue) == False:
colorPos = int(round(colorValue))
rcolor = int(round(cm.jet(colorPos)[0]*255))
gcolor = int(round(cm.jet(colorPos)[1]*255))
bcolor = int(round(cm.jet(colorPos)[2]*255))
rgbcolor = [rcolor, gcolor, bcolor]
thickness = 2
myMaskContoursAllCurvatureValues[int(contoursxy[ii,1])-thickness:int(contoursxy[ii,1])+thickness,int(contoursxy[ii,0])-thickness:int(contoursxy[ii,0])+thickness,:] = rgbcolor
added_image = cv2.addWeighted(myMaskContoursAllCurvatureValues, 0.95, myImageThresholdedRGBConv, 0.01, 0)
frames_all.append(added_image)
frames_all.append(added_image)
frames_all.append(added_image)
if (i == 100):
ret, image = cap.retrieve()
end = time.time()
plt.figure(1)
plt.imshow(added_image)
frame_one = frames_all[0]
imageio.mimsave('./DataExternalMov/35204.gif', frames_all, loop = 0, duration = 0.1)
if saveGallery == True:
imageio.mimsave('./../GALLERY/Analysis_Of_BioImages/03_01_Time_Lapse_Curvature02.gif', frames_all, loop = 0, duration = 0.1)
100.1 .html-Export of Code¶
# export code to html
import os
os.system("jupyter nbconvert AnalysisOfBioImages.ipynb --to html")
fin = open("AnalysisOfBioImages.html", "rt", encoding="utf8")
fout = open("AnalysisOfBioImagesPart01_mod.html", "wt", encoding="utf8")
for line in fin:
if line.find("<ol>") > -1:
line = line.replace("<ol>", "<ol type=\"1\">")
elif line.find("<li>") > -1:
line = line.replace("<li>", "<li type=\"1\">")
fout.write(line)
fin.close()
fout.close()
import os
if os.path.isfile("AnalysisOfBioImages.html"):
os.remove("AnalysisOfBioImages.html")
os.rename("AnalysisOfBioImagesPart01_mod.html", "AnalysisOfBioImages.html")